Abstract
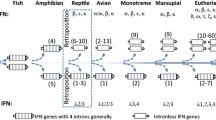
A phylogenetic analysis of mammalian type I interferon (IFN) genes showed: (1) that the three main subfamilies of these genes in mammals (IFN-β, IFN-α, and IFN-ω) diverged after the divergence of birds and mammals but before radiation of the eutherian orders and (2) that IFN-β diverged first. Although apparent cases of interlocus recombination among mouse IFN-α genes were identified, the hypothesis that coding regions of IFN-α genes have been homogenized within species by interlocus recombination was not supported. Flanking regions as well as coding regions of IFN-α were more similar within human and mouse than between these species; and reconstruction of the pattern of nucleotide substitution in IFN-α coding regions of four mammalian species by the maximum parsimony method suggested that parallel substitutions have occurred far more frequently between species than within species. Therefore, it seems likely that IFN-α genes have duplicated independently within different eutherian orders. In general, type I IFN genes are subject to purifying selection, which in the case of IFN-α and IFN-β is strongest in the putative receptor-binding domains. However, analysis of the pattern of nucleotide substitution among IFN-ω genes suggested that positive Darwinian selection may have acted in some cases to diversify members of this subfamily at the amino acid level.
Similar content being viewed by others
References
Adolf GR (1987) Antigenic structure of human interferon ω1 (interferon alpha II): comparison with other human interferons. J Gen Virol 68: 1669–1676
Capon DJ, Shepard HM, Goeddel DV (1985) Two distinct families of human and mouse interferon-α genes are coordinately expressed and encode functional polypeptides. Mol Cell Biol 5:768–779
Carroll RL (1988) Vertebrate paleontology and evolution. WH Freeman, New York
De Maeyer E, De Maeyer-Guignard J (1988) Interferons and other regulatory cytokines. Wiley, New York
Easteal S (1990) The pattern of mammalian evolution and the relative rate of molecular evolution. Genetics 124:165–173
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fish EN (1992) Definition of receptor binding domains in interferon-α. J Interferon Res 12:257–266
Gilliespie D, Carter W (1983) Concerted evolution of human interferon alpha genes. J Interferon Res 3:83–88
Hentschel CC, Birnstiel ML (1981) The organization and expression of histone gene families. Cell 25:301–313
Higgins DG, Bleasby AJ, Fuchs R (1992) Clustal V: improved software for multiple sequence alignment. CABIOS 8:189–191
Hughes AL (1990) Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics 127:345–353
Hughes AL (1992) Positive selection and interallelic recombination at the merozoite surface antigen-1 (MSA-1) locus of Plasmodium falciparum. Mol Biol Evol 9:381–393
Hughes AL (1993) Evidence of positive selection at the Lyb-2 locus of the mouse. Immunogenetics 38:54–56
Hughes AL (1994) The evolution of functionally novel proteins after gene duplication. Proc R Soc Lond [Biol] 256:119–124
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Hughes AL, Nei M (1989) Nucleotide substitution at major histocompafibility complex class II loci: evidence for overdominant selection. Proc Natl Acad Sci USA 86:958–962
Hughes AL, Ota T, Nei M (1990) Positive Darwinian selection promotes charge profile diversity in the antigen binding cleft of class I MHC molecules. Mol Biol Evol 7:515–524
Hughes AL, Hughes MK, Watkins DL (1993) Contrasting roles of interallelic recombination at the HLA-A and HLA-B loci. Genetics 133:669–680
Imakawa K, Anthony RV, Kazemi M, Marotti KR, Polites HG, Roberts RM (1987) interferon-like sequence of ovine trophoblast protein secreted by embryonic trophoectoderm. Nature 330:377–379
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Korn AP, Rose DE, Fish EN (1994) Three-dimensional model of a human interferon-α consensus sequence. J Interferon Res 14:1–9
Li W-H, Gouy M, Sharp PM, O'hUigin C, Yang Y-W (1990) Molecular phylogeny of Rodentia, Lagomorpha, Primates, Artiodactyla, and Carnivora and molecular clocks. Proc Natl Acad Sci USA 87:6703–6707
May LT, Sehgal PB (1985) On the relationship between human interferon α1 and β1 genes. J Interferon Res 5:521–526
Miyata T, Hayashida H (1982) Recent divergence from a common ancestor of human IFN-α genes. Nature 295:165–168
Mouchel-Vielh E, Lutfalla G, Mogensen KE, Uze G (1992) Specific antiviral activities of the human a interferons are determined at the level of receptor (IFNAR) structure. FEBS Lett 313:255–259
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nei M, Jin L (1989) Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol 6: 290–300
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425
Senda T, Shimazu T, Matsuda S, Kawano G, Shimuzu H, Nakamura KT, Mitsui Y (1992) Three-dimensional crystal structure of recombinant murine interferon-β. EMBO J 11:3193–3201
Shaw GD, Boll W, Taiva H, Mantel N, Lengyel P, Weissmann C (1983) Structure and expression of cloned murine IFN-α genes. Nucleic Acids Res 11:555–572
Stephens JC (1985) Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol 2:539–556
Swofford DL (1990) PAUP: phylogenetic analysis using parsimony. Version 3.0. Illinois Natural History Survey, Champaign
Tamai T, Sirahata S, Noguchi T, Sato N, Kimura S, Hiroki M (1993) Cloning and expression of flatfish (Paralichrhys olivaceus) interferon cDNA. Biochim Biophys Acta 1174:182–186
Todokoro K, Kioussis D, Weissman C (1984) Two non-allelic human interferon genes with identical coding regions. EMBO J 3:1809–1812
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hughes, A.L. The evolution of the type I interferon gene family in mammals. J Mol Evol 41, 539–548 (1995). https://doi.org/10.1007/BF00175811
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00175811