Abstract
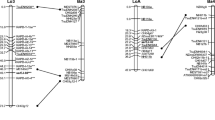
A genetic linkage map of Prunus has been constructed using an interspecific F2 population generated from self-pollinating a single F1 plant from a cross between a dwarf peach selection (54P455) and an almond cultivar ‘Padre’. Mendelian segregations were observed for 118 markers including 1 morphological (dw), 6 isozymes, 12 plum genomic, 14 almond genomic and 75 peach mesocarp specific cDNA markers. One hundred and seven markers were mapped to 9 different linkage groups covering about 800 cM map distance, and 11 markers remained unlinked. Three loci identified by three cDNA clones, PC8, PC5 and PC68.1, were tightly linked to the dw locus in linkage group 5. Segregation distortion was observed for approximately one-third of the markers, perhaps due to the interspecific nature and the reproductive (i.e. self-incompatibility) differences between peach and almond. This map will be used for adding other markers and genes controlling important traits, identifying the genomic locations and genetic characterizing of the economically important genes in the genus Prunus, as well as for markerassisted selection in breeding populations. Of particular interest are the genes controlling tree growth and form, and fruit ripening and mesocarp development in peach and almond.
Similar content being viewed by others
References
Arulsekar S, Parfitt DE (1986) Isozyme analysis procedures for stone fruits, almond, grape, walnut and fig. HortScience 21:928–933
Arulsekar S, Parfitt DE, Kester DE (1986) Comparison of isozyme variability in peach and almond cultivars. J Hered 77:272–274
Arumuganathan K, Earle E (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Arus P, Olarte C, Romero M, Vargas F (1994) Linkage analysis of ten isozyme genes in F1 segregating almond progenies. J Am Soc Hort Sci 119:339–344
Belthoff LE, Ballard R, Abbott A, Baird WV,Morgens P, Callahan A, Scorza R, Monet R (1993) Development of a saturated linkage map of Prunus persica using molecular based marker systems. Acta Hortic 336:51–56
Bernatzky R, Tanksley SD (1986) Towards a saturated linkage map in tomato based on isozymes and random cDNA sequences. Genetics 112:887–898
Chaparro JX, Werner DJ, O'Malley DO, Sederoff RR (1994) Targeted mapping and linkage analysis of morphological, isozyme and RAPD markers in peach. Theor Appl Genet 87:805–815
Durham RE, Liou PC, Gmitter FC Jr, Moore GA (1992) Linkage of restriction fragment length polymorphisms and isozymes in Citrus. Theor Appl Genet 84:39–48
Feinberg AP, Vogelstein B (1983) A technique for radiolabelling fragments to high specific activity. Anal Biochem 132:6–13
Gebhardt C, Ritter E, Barone A, Debener T, Walkemeier B, Schachtschabel U, Kaufmann H, Thompson R, Bonierbale M, Ganal M, Tanksley S, Salamini F (1991) RFLP maps of potato and their alignment with the homoeologous tomato genome. Theor Appl Genet 83:49–57
Gepts P, Clegg MT (1989) Genetic diversity in pearl millet (Pennisetum glaucum) at the DNA sequence level. J Hered 80:203–208
Haldane JBS (1919) The combination of linkage values and the calculation of distances between the loci of linked factors. J Genet 8:299–309
Hesse CO (1975) Peach. In: Janick J, Moore J (eds) Advances in fruit breeding. Purdue University Press, West Lafayette, Ind., pp285–335
Jarrell DC, Roose ML, Traugh SN, Kupper RS (1992) A genetic map of citrus based on the segregation of isozymes and RFLPs in an in intergeneric gross. Theor Appl Genet 84:49–56.
Kester DE, Asay RN (1975) Almond. In: Janick J, Moore J (eds) Advances in fruit breeding. Purdue University Press, West Lafayette, Ind., pp 387–419
Lander ES, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newberg L (1987) MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lincoln S, Daly M, Lander E (1992) Constructing genetic maps with MAPMAKER/EXP 3.0. Whitehead Institute Technical Report. 3rd edn., Cambridge, Mass.
Messeguer R, Viruel MA, de Vicente MC, Garcia-Mas J, Fernandez-Busquets X, Vargas F, Puigdomenech P, Arus P (1994) Construction of a genetic map with molecular markers in almond. Plant Genome III:51 (abstr)
Mowrey B, Werner D, Byrne D (1990) Inheritance of isocitrate dehydrogenase, malate dehydrogenase and shikimate dehydrogenase in peach and peach almond hybrids. J Am Soc Hortic Sci 115:312–319
Nienhuis J, Helentjaris T, Slocum M, Ruggero R, Schaefer A (1987) Restriction fragment length polymorphism analysis of loci associated with insect resistance in tomato. Crop Sci 27:797–803
Paterson AH, Tanksley SD, Sorrells ME (1991) DNA markers in plant improvement. Adv Agron 46:39–90
Rajapakse S, Belthoff LE, Scorza R, Ballard RE, Baird WV, Monet R, Abbott AG (1994) Genetic linkage mapping peach using morphological, RFLP, and RAPD markers. Plant Genome III:56(abstr)
Ritter E, Gebhardt C, Salamini F (1990) Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 125: 645–654
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Rihosomal DNA spaces-length polymorphism in barley Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Scorza R, Mehlenbacher SA, Lighner GW (1985) Inbreeding and coancestry of freestone peach cultivars of the eastern United States and implications for peach germplasm improvement. J Am Soc Hortic Sci 110:547–552
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98: 503–517
Tanksley SD, Young ND, Paterson AH, Bonierbale MW (1989) RFLP Mapping in plant breeding: New tools for an old science. Bio/technology 7:257–264
Weeden NF (1989) Application of isozymes in plant breeding. In: Janick J (ed) Plant breeding reviews, vol 6. Timber Press, Portland, Ore., pp 11–54
Whitkus R, Doebley J, Lee M (1992) Comparative genome mapping of sorghum and maize. Genetics 132:1119–1130
Zamir D, Tadmor Y (1986) Unequal segregation of nuclear genes in plants. Bot Gaz 147:355–358
Author information
Authors and Affiliations
Additional information
Communicated by A. L. Kahler
Rights and permissions
About this article
Cite this article
Foolad, M.R., Arulsekar, S., Becerra, V. et al. A genetic map of Prunus based on an interspecific cross between peach and almond. Theoret. Appl. Genetics 91, 262–269 (1995). https://doi.org/10.1007/BF00220887
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00220887