Summary
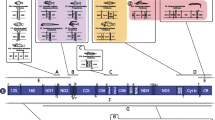
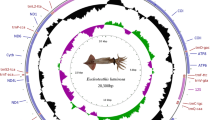
The gene organization of starfish mitochondrial DNA is identical with that of the sea urchin counterpart except for a reported inversion of an approximately 4.6-kb segment containing two structural genes for NADH dehydrogenase subunits 1 and 2 (ND 1 and ND 2). When the codon usage of each structural gene in starfish, sea urchin, and vertebrate mitochondrial DNAs is examined, it is striking that codons ending in T and G are preferentially used more for heavy strand-encoded genes, including starfish ND 1 and ND 2, than for light strand-encoded genes, including sea urchin ND 1 and ND 2. On the contrary, codons ending in A and Care preferentially used for the light strand-encoded genes rather than for the heavy strand-encoded ones. Moreover, G-U base pairs are more frequently found in the possible secondary structures of heavy strandencoded tRNAs than in those of light strand-encoded tRNAs. These observations suggest the existence of a certain constraint operating on mitochondrial genomes from various animal phyla, which results in the accumulation of G and T on one strand, and A and C on the other.
Similar content being viewed by others
References
Aloni Y, Attardi G (1971) Expression of the mitochondrial genome in Hela cells. II. Evidence for complete transcription of mitochondrial DNA. J Mol Biol 55:251–270
Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Anderson S, de Bruijn MHL, Coulson AR, Eperon IC, Sanger F, Young IG (1982) Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol 156:683–717
Bibb MJ, van Etten RA, Wright CT, Walberg MW, Clayton DA (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell 26:167–180
Bogenhagen D, Clayton DA (1978) Mechanism of mitochondrial DNA replication in mouse L-cells: kinetics of synthesis and turnover of the initiation sequence. J Mol Biol 119:49–68
Bolden A, Noy GP, Weissbach A (1977) DNA polymerase of mitochondria is a γ-polymerase. J Biol Chem 252:3351–3356
Brown WM (1981) Mechanisms of evolution in animal mitochondrial DNA. Ann NY Acad Sci 361:119–134
Brown WM, Prager EM, Wang A, Wilson AC (1982) Mitochondrial DNA sequence of primates: tempo and mode of evolution. J Mol Evol 18:225–239
Cantatore P, Roberti M, Rainaldi G, Gadaleta MN, Saccone C (1989) The complete nucleotide sequence, gene organization, and genetic code of mitochondrial genome ofParacentrotus lividus. J Biol Chem 264:10965–10975
Chang DD, Clayton DA (1985) Priming of human mitochondrial DNA replication occurs at the light-strand promoter. Proc Natl Acad Sci USA 82:351–355
Clary DO, Wolstenholme DR (1985) The mitochondrial DNA molecule ofDrosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 22:252–271
Clayton DA, Doda JN, Friedberg EC (1974) The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci USA 71:2777–2781
Desjardins P, Morais R (1990) Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher vertebrates. J Mol Biol 212:599–634
Doda JN, Wright CT, Clayton DA (1981) Elongation of displacement-loop strands in human and mouse mitochondrial DNA is arrested near specific template sequences. Proc Natl Acad Sci USA 78:6116–6120
Gadaleta G, Pepe G, De Candia G, Quagliariello C, Sbisá E, Saccone C (1989) The complete nucleotide sequence of theRattus norvegicus mitochondrial genome: cryptic signals revealed by comparative analysis between vertebrates. J Mol Evol 28:497–516
Goddard JM, Wolstenholme DR (1980) Origin and direction of replication in mitochondrial DNA molecules from the genusDrosophila. Nucleic Acids Res 8:741–757
Hasegawa M, Kishino H (1989) Heterogeneity of tempo and mode of mitochondrial DNA evolution among mammalian orders. Jpn J Genet 64:243–258
Heckman JE, Sarnoff J, Alzner-De Weed B, Yin S, RajBhandary UL (1980) Novel features in the genetic code and codon reading patterns inNeurospora crassa mitochondria based on sequences of six mitochondrial tRNAs. Proc Natl Acad Sci USA 77:3159–3163
Himeno H, Masaki H, Kawai T, Ohta T, Kumagai I, Miura K, Watanabe K (1987) Unusual genetic codes and a novel gene structure for tRNA SerAGY in starfish mitochondrial DNA. Gene 56:219–230
Ikemura T (1981) Correlation between the abundance ofEscherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for theE. coli translational system. J Mol Biol 151:389–409
Ikemura T (1982) Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. J Mol Biol 158:573–597
Jacobs HT, Elliott DJ, Math VB, Farquharson A (1988) Nucleotide sequence and gene organization of sea urchin mitochondrial DNA. J Mol Biol 202:185–217
Jacobs HT, Asakawa S, Araki T, Miura K, Smith MJ, Watanabe K (1989a) Conserved tRNA cluster in starfish mitochondrial DNA. Curr Genet 15:193–206
Jacobs HT, Herbert ER, Rankine J (1989b) Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication orgigin region. Nucleic Acids Res 17: 8949–8965
Jukes TH, Bhushan V (1986) Silent nucleotide substitutions and G+C content of some mitochondrial and bacterial genes. J Mol Evol 24:39–44
Jukes TH, Osawa S, Muto A, Lehman N (1987) Evolution of anticodons: variations in the genetic code. Cold Spring Harbor Symp Quant Biol 52:769–776
Kumazawa Y, Yokogawa T, Hasegawa E, Miura K, Watanabe K (1989) The aminoacylation of structually variant phenylalanine tRNAs from mitochondria and various nonmitochondria sources by bovine mitochondrial phenylalanyl-tRNA synthetase. J Biol Chem 264:13005–13011
Kunkel TA (1985) The mutational specificity of DNA polymerase-α and-γ duringin vitro DNA synthesis. J Biol Chem 260:12866–12874
Martens PA, Clayton DA (1979) Mechanism of mitochondrial DNA replication in mouse L-cell: localization and sequence of the light-strand origin of replication. J Mol Biol 135:327–351
Muto A, Osawa S (1987) The guanine and cytosine content of genomic DNA and bacteria evolution. Proc Natl Acad Sci USA 84:166–169
Ohama T, Muto A, Osawa S (1990) Role of GC-biases mutation pressure on synonymous codon choice inMicroccus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res 18:1565–1569
Osawa S, Jukes TH (1989) Codon reassignment (codon capture) in evolution. J Mol Evol 28:271–278
Osawa S, Ohama T, Yamao F, Muto A, Jukes TH, Ozeki H, Umesono K (1988) Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc Natl Acad Sci USA 85:1124–1128
Robberson DL, Kasamatsu H, Vinograd J (1972) Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc Natl Acad Sci USA 69:737–741
Roe BA, Wong JFH, Chen EY, Armstrong PA (1981) Sequence analysis of mammalian mitochondrial tRNAs. In: Walton AG (ed) Proceedings of the third Cleveland symposium. Elsevier, Amsterdam, pp 167–176
Roe BA, Ma D-P, Wilson RK, Wong JFH (1985) The complete nucleotide sequence of theXenopus laevis mitochondrial genome. J Biol Chem 260:9759–9774
Smith MJ, Banfield DK, Doteval K, Gorski S, Kowbel DJ (1989) Gene arrangement in sea star mitochondrial DNA demonstrates a major inversion event during echinoderm evolution. Gene 76:181–185
Sprinzl M, Hartmann, T, Weber J, Blank J, Zeidler R (1989) compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 17:r1-r171
Sueoka N (1962) On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci USA 48:582–592
Takeishi K, Gotoh O (1982) Sequence relationship among various 4.5S RNA species. J Biochem 92:1173–1177
Thomas WK, Maa J, Wilson AC (1989) Shifting constraints on tRNA genes during mitochondrial DNA evolution in animals. New Biol 1:93–100
Wolstenholme DR, Macfarlane JL, Okimoto R, Clary DO, Wahleithner JA (1987) Bizarre tRNAs inferred from DNA sequences of mitochondrial genomes of nematode worms. Proc Natl Acad Sci USA 84:1324–1328
Yokogawa T, Kumazawa Y, Miura K, Watanabe K (1989) Purification and characterization of two serine isoaccepter tRNAs from bovine mitochondria by using a hybridization assay method. Nucleic Acids Res 17:2623–2638
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Asakawa, S., Kumazawa, Y., Araki, T. et al. Strand-specific nucleotide composition bias in echinoderm and vertebrate mitochondrial genomes. J Mol Evol 32, 511–520 (1991). https://doi.org/10.1007/BF02102653
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02102653