Abstract
The mitotic spindle uses dynamic microtubules and mitotic motors to generate the pico-Newton scale forces that are needed to drive the mitotic movements that underlie chromosome capture, alignment and segregation. Here, we consider the biophysical and molecular basis of force-generation for chromosome movements in the spindle, and, with reference to the Drosophila embryo mitotic spindle, we briefly discuss how mathematical modeling can complement experimental analysis to illuminate the mechanisms of chromosome-to-pole motility during anaphase A and spindle elongation during anaphase B.
Similar content being viewed by others
Introduction
Mitosis is the process by which identical sets of genetic instructions are delivered to the product of every cell division (Fig. 1). This process was first described in detail and illustrated by Walter Fleming more than a century ago [1], and it is now understood to be coordinated by the mitotic spindle (Fig. 1a), a subcellular machine which uses microtubules (MTs) and multiple mitotic motors to assemble itself and to accurately segregate the genome into the daughter nuclei. Errors in this process can have devastating consequences for the cell and the organism since they can lead to genomic instability, birth defects and cancer.
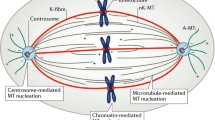
The mitotic spindle. a Three different classes of MTs make up the mitotic spindle, namely astral aMTs, interpolar ipMTs and kinetochore kMTs. b Chromosome motility during different phases of mitosis; chromosomes are captured during prometaphase (upper), aligned at the spindle equator during metaphase (center) and segregated by chromosome-to-pole motion during anaphase A, plus spindle elongation during anaphase B (lower). c Schematic diagram of the experimental set-up that uses calibrated microneedles to measure the force exerted by the mitotic spindle on an anaphase chromosome (upper) compared with estimated force required to move a chromosome against viscous drag (lower)
Mitosis proceeds in distinct phases. In astral spindles, which assemble predominantly via a centrosome-controlled pathway, dynamic MTs of the bipolar spindle capture chromosomes, each consisting of pairs of sister chromatids, during prometaphase (Fig. 1b). By metaphase, captured sister chromatids are aligned at the spindle equator, and a constant spindle length is maintained for a period that can last from seconds to years in different types of spindles. Subsequently, the sister chromatids segregate as a result of being pulled polewards by disassembling kinetochore fibers during anaphase A, and also by the elongation of the spindle itself, which increases pole–pole spacing, during anaphase B (Fig. 1b). Finally, the daughter nuclei form around the segregated and decondensing chromatin while the spindle is disassembled during telophase.
A mitotic spindle displays a reasonably predictable degree of structural order; spindle MTs are extremely dynamic and are generally organized with their minus ends at or near the spindle poles with their plus ends extending outward to form three sets, the astral (aMT), interpolar (ipMT) and kinetochore (kMT) microtubules (MTs) (Fig. 1a). These MTs can potentially serve as polymer ratchets to exert forces that push and pull the chromosomes or the spindle poles, and as dynamic, polarized tracks for the force-generating mitotic motors that move cargo along them; for example, a chromosome, a cell cycle (or other) regulatory molecule, or a second “cargo” MT that is crosslinked by the motor to its MT track (see below). In this review, we focus on force generation for chromosome movement (see [2] for an earlier, comprehensive review of this topic). Force generation for spindle assembly and length control has been recently reviewed elsewhere [3].
Force, work and energy underlying chromosome motility in the mitotic spindle
In the sub-cellular regime, low Reynolds number conditions dominate motility (i.e. Re < 10−3, where \({Re} = \frac{\text{inertial\, drag}}{\text{viscous\, drag}} = \frac{vL\rho }{\eta } \)), so that inertial forces (due to mass) are negligible in driving movements, and viscous forces (due to friction) have the major effect [4, 5]. For example, in Drosophila syncytial embryo spindles, anaphase A lasts about ~50 s during which sister chromatids move from the equator towards the opposite poles at a rapid rate of v = 0.1 μm s−1 while the spindle is maintained at a constant length of 12 μm. A fly chromosome of length L ~ 3 μm and density ρ = 103 kg m−3, moving through cytoplasm of viscosity ~0.2 pN s μm−2 (see below), will have Reynolds number, Re ~ 10−9 ≪ 10−3. This low value for the Reynolds number means that the chromosomes’ movement is dominated by viscous forces, and hence the net force driving the movement of the chromosome is proportional to its velocity and its viscous drag coefficient: F = μv (Fig. 1c).
It is possible to estimate the viscous drag coefficient of a chromosome, based on its size and shape, and the estimated cytoplasmic viscosity [5]. The viscous drag coefficient associated with a fly chromosome has been estimated using in vivo data [6]. In this study, the Young’s modulus of the chromosome was determined based on the 4D analysis of its motion (curvature fluctuations), and using this value, the net poleward force powering its movement during anaphase A was estimated, and found to be of the order of 0.1–1 pN. This net force value, combined with the observed anaphase A rate, yielded an estimate for the drag coefficient of a chromosome and also for the nucleoplasmic viscosity.
The estimate of the drag force acting during chromosome-to-pole movement can be used to calculate the work done and the energy used in moving a fly chromosome from the equator to the pole (Work = Force × Distance ~ 10−12N × 10−5 m = 10−17 J). ATP hydrolysis releases about 50 kJ mole−1 free energy under cellular conditions [5], so a single ATP molecule yields ΔG ≈ 50 × 103/(6 × 1023) = 10−19 J. Therefore, assuming (conservatively; see below) that the force generators (MTs or motors) have only ~10% coupling efficiency, as few as 1,000 ATP hydrolyses are sufficient to move a fly chromosome a distance of the order of 10 μm from the equator to the pole at the observed rapid rate of 0.1 μm s−1 against viscous drag. Since a typical dividing cell consumes 107 ATP per second [5], this amounts to only a minute fraction of the cell’s energy.
Forces measured for moving chromosomes in the mitotic spindle
Chromosome-to-pole motility
How much poleward force can the mitotic force generators, namely, MT polymer ratchets and kinetochore motors exert on a chromosome, e.g., during prometaphase and anaphase (Fig. 1b)? In one important study, Alexander and Rieder [7] measured the net force powering the rapid poleward movements of attaching chromosomes during prometaphase in Newt lung cells, and estimated ~1–10 pN force per MT. In classic, pioneering experiments, Bruce Nicklas measured the maximal force that can be exerted by the spindle on a chromosome during anaphase A in grasshopper spermatocytes [8]. Using force-calibrated flexible glass needles, he snagged chromosomes during anaphase A, and measured forces of the order of 700 pN being required to stall chromosome to pole movement (Fig. 1c). A most intriguing aspect of Nicklas’ results was that the chromosome did not slow down until forces of 200–300 pN were applied to it.
This latter result remains somewhat mysterious, in view of the estimate that only ~1 pN scale forces are required to move the chromosome poleward during anaphase A. Why does a chromosome use so little of the spindle’s force-generating capacity, and how does the chromosome’s velocity remain constant when challenged by an opposing force corresponding to a 100-fold increase in load? Nicklas suggested that this was an indication of the presence of a ‘governor’ for motility, such as the kMT. For example, the kMT could act as a barrier to poleward motion so its loss by depolymerization rather than the kinetochore motor’s motility rate limits the chromosome’s velocity. In this view, assuming that there are ~15 kMTs per kinetochore, as few as ~5–10 motors per MT-kinetochore interface tracking the plus end of the kMT could produce the large force Nicklas observed, without a significant slowing down of the snagged chromosome. That is, 15 × 10 = 150 motors would be stalled by a force in the range of ~1000 pN. Since chromatid to pole velocity is limited by the shortening of the k-fiber, an opposing load would not slow down the kinetochore motors until a force of ~500 pN is applied. However, in the absence of such an external load, a single attached motor (e.g., a single dynein) or a single depolymerizing MT is sufficient to drive the observed chromosome to pole motility against viscous drag. The elucidation of the mechanistic basis of the discrepancy between the level of force powering the chromosomes polewards in the absence and the presence of an external load will require improved understanding of the dynamic coupling between the kinetochore and the kMT, the identity of the force-generator(s), and a careful structural analysis of the kinetochore-MT interface during anaphase. Even though progress in these areas has been achieved, it remains an active topic of research.
Polar ejection forces
In the Drosophila embryo spindle, the magnitude of the polar ejection forces (PE), i.e. the forces generated by polymerizing MTs and chromosome arm-associated kinesins (below) that push the chromosome arms away from the poles during prometaphase and metaphase (Fig. 1b), was estimated to be ~1 pN per MT, based on the computational analysis of 4D chromosome motion and modeling [9]. In agreement with Nicklas’ measurements, these studies also suggest that the magnitude of the maximal force exerted on the chromosomes by spindle MTs and motors lies in the range of several hundreds to thousands of pN.
Recent studies have examined the spatial distribution of these polar ejection forces by amputation of the chromosome arm in order to reduce PE forces while leaving the kinetochore motility machinery intact, and then tracking the oscillations of the remaining kinetochore–arm complex in the spindle [10]. Arm amputation was observed to increase the amplitude of the oscillations without altering the poleward and anti-poleward velocity of the kinetochores. The position at which the oscillating chromosomes (intact or with an amputated arm) reverse direction is assumed to correspond to the maximal PE force the kinetochore machinery can withstand. Based on this, the experimental observations could be fit to a PE force-position function in the spindle, where the exponent of the position was treated as a free parameter. Contrary to the current view that MT plus ends are exponentially distributed so that PE forces decrease strongly away from the pole (as an inverse square function), the results suggest that the PE force is relatively high but constant throughout a surprisingly wide region moving from the poles and decreases only linearly towards the equatorial region of the spindle [10]. This finding draws attention to the need for further investigation of the distribution of MT plus ends and motors in the metaphase spindles that keep the chromosomes aligned at the metaphase plate despite their short poleward and anti-poleward excursions.
Structural organization of spindle MTs
Structural studies have revealed that MTs, the major sites of force-generation for chromosome movement in the spindle, are 13 stranded polymers, in which ~8-nm-long αβ-tubulin dimers are polymerized end to end with the α subunits facing the MT’s minus end (Figs. 2a and 3a) [11–13]. This structural polarity is important since it gives rise to the different polymerization/depolymerization kinetics of the MT plus and minus ends, and it controls the directional motility of motors that move relative to the polymer lattice. For example, the MT plus ends exhibit more ‘active’ dynamic instability (DI) than the minus ends in vitro (Fig. 2a), whereas some mitotic motors, like kinesin-5, move towards the MT plus ends while others such as kinesin-14 and cytoplasmic dynein move towards their minus ends (Fig. 3b).
Microtubule structure and dynamics in the spindle. a Left panel MT dynamic instability (DI) observed in vitro and plots of the associated MT length changes. Right panel turnover dynamics of MTs in the spindle assayed by fluorescence recovery after photobleaching (FRAP) where the rapid recovery is best explained by DI of spindle MTs. b Schematic representation of the use of serial section EM of a mitotic spindle at metaphase: 3D organization and position of the MTs and chromosomes in the spindle is reconstructed based on the analysis of multiple thin cross-sections of which only two are shown. Spindle MT structure in three dimensions is reconstructed by tracking MT trajectories from section to section. c Poleward flux of spindle MTs as observed by fluorescence speckle microscopy (FSM). Example taken from Drosophila embryos where small groups of tubulin dimers in individual ipMT bundles are observed to move away from the spindle equator from metaphase through anaphase B (oblique traces of speckles are drawn superimposed on the right hand kymograph). During metaphase and anaphase A (termed pre-anaphase B), when pole–pole distance is maintained at a constant spacing, tubulin speckles are observed to move towards the poles, in the process termed poleward flux. During anaphase B, speckles move away from the equator at the same rate as the poles as the spindle elongates, pulling the chromosomes, together with the poles, away from the equator. Lower diagram illustrates a model for poleward flux of ipMTs during pre-anaphase B and the persistent movement of speckles away from the equator at the same rate as the poles during anaphase B
Force-generators in the mitotic spindle. a Nucleotide hydrolysis by MTs and MT-based motors is used for polymer dynamics and to bias thermal fluctuations to generate directional force and motility by polymer ratchets (left) and mitotic motors (right). Only a single MT protofilament is shown. b Cartoon showing hypothetical coupling between the chemical and mechanical changes associated with directional stepping of a two-headed motor along a polymer. ATP hydrolysis releases free energy that biases the directionality of conformational changes to occur in the sequence 1-to-2-to-3-to-1, since the reverse sequence of conformational changes is coupled to ATP synthesis and is therefore of low probability. On the reaction coordinate diagram (below), ATP hydrolysis tilts the energy landscape downward to the right so that the forward rate constant (K f) exceeds the reverse rate constant (K r) and consequently the motor tends to step persistently rightward. An increasing opposing force (F load) tends to antagonize the ATP-induced downward tilt, causing the motor to slow down in accordance with its force–velocity profile until K f = K r at F stall, whereupon the motor stops. Assuming thermodynamic equilibrium between the pre-step (n) and post-step (n +1) states, the motor can do work W = Fδ, and from the Boltzman distribution \( F_{\text{eq}} = \,{\frac{{k_{\text{B} } T}}{\delta }}\ln K_{\text{eq}} \). c Force-generation by the rectification of thermal energy via the action of MTs working as ‘Brownian Ratchets’. As shown in the reaction coordinate diagram, the presence of a load (e.g., a chromosome) ‘tilts’ the energy landscape, so that the free energy released, i.e. the binding energy, ΔG B, (due to the formation of inter-subunit weak bonds as tubulin polymerizes), is reduced by Fδ (=work done to push the chromosome/load a distance δ)
The organization and polarity pattern of MTs in the spindle was determined using elegant systematic electron microscopic studies by McIntosh et al. [14–18]. The 3D reconstruction of MT trajectories from serial EM sections, combined with polarity assays by hook decoration, revealed several key features of the spindle MTs’ spatial and structural organization (Fig. 2b). It was this work which determined that many spindles contain three classes of MTs (Fig. 1a): (1) kinetochore MTs (kMTs) that link the chromosomes to the poles, (2) the interpolar MTs (ipMTs) that emanate from the poles towards and beyond the spindle equator, and (3) astral MTs (aMTs) that extend outward from the centrosomes. In vertebrate tissue culture cells, for example, it was observed that the kMTs have their minus ends all facing the spindle poles and run all the way from the kinetochores to the poles. The latter result is perhaps surprising, given the subsequent discovery of MT nucleation along the walls of pre-existing MTs within the spindle (e.g., by the augmin complex) [19]. In contrast, it was observed that the trajectories of MTs within ipMT bundles are less persistent than those of kMTs, often having their minus ends detached from the centrosome at the pole and sometimes being associated with kMT bundles instead. The plus ends of some of these ipMTs overlap in an anti-parallel orientation around the spindle equator (midzone). Both the kMTs and ipMTs are found in bundles, possibly being crosslinked by motors and non-motor MT-associated proteins (MAPs) to form a mechanically inter-connected continuum.
Dynamics of spindle MTs
MTs in living mitotic spindles were initially visualized using polarized light microscopy [20], which documented the rapid polymerization/depolymerization dynamics of spindle MTs. In the early 1980s and 1990s, newly developed imaging techniques combined with the use of fluorescently-tagged tubulin further revealed the dynamics of MTs in live spindles [21–23]. For example, fluorescence recovery after photobleaching (FRAP) revealed that spindle MTs in cultured mammalian cells turn-over rapidly with a half life of ~20 s, which could be explained by the dynamic instability of MT plus ends following first order kinetics (Fig. 2a) [22]. In complementary studies, cold treatment of mitotic cells selectively eliminated the non-kMTs, and it was revealed that the kMTs turn over less rapidly (half life–several minutes) than the non-kMTs [24, 25].
Also, in elegant studies, the persistent poleward movement of tubulin dimers within the spindle MTs of tissue culture cells was observed by the photoactivation of caged-fluorescent tubulin incorporated into mitotic spindles [23]. This “poleward flux” can be explained both at the molecular and theoretical level by the poleward sliding of MTs coupled to the depolymerization of their minus ends at the spindle poles and incorporation of new tubulin subunits at kinetochores [26, 27] (e.g., Fig. 2c). However, based on studies of poleward flux in Xenopus egg extract spindles, an alternative “slide-and-cluster” model has been proposed [28]. The development of Fluorescence Speckle Microscopy (FSM) and its application to studying MT dynamics in the mitotic spindles allowed the visualization of the dynamics of groups of tubulin dimers within spindle MTs in various organisms [29–32]. With the notable exception of the budding yeast, it was observed that MTs of all spindles examined undergo poleward flux at rates ranging from 0.005 to 0.06 μm s−1 and the non-kMTs of metaphase spindles turnover rapidly, with a half life ranging from a few seconds to a few minutes. Naturally, these findings led to the question of how the mitotic spindle maintains a robust structure capable of segregating the genome as accurately as it does, despite the constant turnover and poleward movement of MTs that make up the spindle and capture and segregate the chromosomes [33].
Molecular mechanisms of force generation for chromosome movements
The idea that the polymerization and depolymerization of spindle fibers generate forces in the spindle was a cornerstone of Shinya Inoue’s ‘Dynamic Equilibrium Model’ for mitosis, in which he postulated that spindle fibers act like liquid crystals that grow/polymerize to push the poles apart and shrink/depolymerize to pull the chromosomes to the poles [34].
Following the discovery of tubulin and its ability to self-assemble in vitro [35–37], it became clear that the spindle fibers are made of MTs, dynamic cytoskeletal polymers whose role in force-generation is quite well understood [38]. They can polymerize to exert pushing (compressive) forces and depolymerize to exert pulling (tensile) forces on the chromosomes and the spindle poles. It is further understood that the dynamic instability (DI) of 13-strand MT plus ends is characterized by stochastic transitions (catastophes and rescues occurring at frequencies, f cat and f res) between phases of growth (polymerization rate, V g) and shortening (depolymerization rate, V s) [39] (Figs. 2a and 3a, c). The free energy released by intersubunit bond formation during polymerization and GTP hydrolysis during depolymerization can be harnessed to push and pull the chromosomes and the spindle poles, thereby driving mitotic movements. It is thought that a single polymerizing or depolymerizing MT can generate 10–100 pN force in vitro [40, 41], and consequently tens of MTs can cooperate to generate nN scale forces in the spindle. However, many problems remain: for example, how does a depolymerizing MT tip remain attached to its cargo?
A large body of evidence suggests that MT polymer ratchets cooperate with kinesin and dynein motors (Fig. 3b, c) to generate the forces that drive motility during mitosis. There are multiple MT-based motors in the spindle, including plus and minus end-directed kinesins and dynein, each capable of generating ~1–10 pN force per motor in vitro [42, 43]. For example, in biophysical studies, the mitotic motor, kinesin-5 (below), was observed to take 8 nm steps (δ), and typically generated forces of 4 pN (although values as high as 7 pN were observed) [43]. The maximum force such a motor could exert by hydrolyzing one ATP per step (step size = δ), is given by F max = energy (ΔG ATP)/distance (δ), i.e. 80 J/8 nm = 10 pN, which suggests a coupling efficiency of greater than 40%.
The magnitude of forces observed suggests that, in principle, multiple motors and MT polymer ratchets could cooperate or compete in the spindle to generate pN–nN scale forces, in order to position and segregate the duplicated chromosomes during mitosis. The basis for both MT- and motor-based force-generation is the rectification of thermal energy (Fig. 3a–c). For example, in the absence of ATP, motors bound to MTs undergo a one-dimensional (1D) random walk using thermal energy, but an ordered series of conformational changes coupled to sequential steps in the ATP hydrolysis reaction renders this movement unidirectional, since the probability of moving backward, thereby synthesizing ATP, is very low (Fig. 3b, upper). On a reaction coordinate diagram (Fig. 3b, lower), it can be seen that ATP hydrolysis “tilts” the motor’s energy landscape so that the motor tends to move to the right because forward steps are more probable than backward steps, i.e. the forward rate constant, k f, from subunit n to (n + 1) is associated with a lower activation energy than the reverse rate constant, from subunit n to (n − 1), but in the absence of ATP hydrolysis, k f = k r, i.e. the forward and backward steps are of equal probability. An opposing force (F load) negates the tilt created by ATP hydrolysis, slowing down the motor’s rightward motion in accordance with its force–velocity curve, until the motor stalls at F stall.
Similarly, MT growth occurs by the binding of GTP-bound tubulin subunits to its tip yielding a supply of free energy due to the formation of noncovalent bonds between tubulin subunits, termed the binding energy (ΔG B) (Fig. 3c). Since the free energy released by this binding exceeds thermal energy (k B T ~ 4.1 pN nm), polymerization is unlikely to be spontaneously reversed, so it rectifies thermal movements of both the MT tip itself and of an object located at the tip. In other words, the binding energy can be used to exert a pushing force (F) that does work (W) on a load to move it a distance δ, where F = W/δ. Conversely, since GTP is hydrolyzed as new tubulin subunits are incorporated into the wall of the MT, part of the free energy released from this hydrolysis is trapped in the MT lattice as stored elastic energy which can subsequently be released during MT depolymerization and used to exert a pulling force on, e.g., a chromosome. Thus, GTP hydrolysis allows a MT polymer ratchet to “pull” as well as to “push”.
Mitotic motors
Discovery of mitotic motors
Biochemical approaches
The first identified MT-based motor, the dynein ATPase, was isolated from cilia in 1965 as a potential MT–MT sliding motor driving ciliary motility [44]. Seminal work on fast axonal transport led to the purification of a second type of motor, kinesin-1, from squid axons [45, 46]. It was shown that kinesin-1 is a MT-plus end-directed motor, moving at ≈1 μm s−1 in the absence of load. Cytoplasmic dynein was purified a couple of years later [47, 48] and shown to move at the same rapid rate of ≈1 μm s−1, but towards the MT-minus ends [49].
In 1969, McIntosh et al. [50] proposed that such MT–MT crosslinking motors could drive a “Sliding Filament Mechanism for Mitosis”. Later, in the early to mid-1980s many laboratories, using MT-affinity, isolated ATPases that might generate forces for intracellular movements including MT–MT sliding and chromosome movement in the spindle. For example, using MT affinity purification, ATPase and motility assays, and pan-kinesin peptide antibody screens, several putative mitotic motors were identified from early echinoderm and Drosophila embryos [51–54]. One of these turned out to be KLP61F, a member of the kinesin-5 family, which was purified from Drosophila embryos as a slow MT-based motor, moving towards the MT plus end at 0.04 μm s−1, with a bipolar homotetrametic structure consisting of pairs of motor heads at opposite ends of a four-strand coil–coil rod [55]. Based on this ultrastructure and the corresponding mutant phenotype (below), it was proposed that kinesin-5 motors such as KLP61F could crosslink and slide anti-parallel overlapping MTs, for example, driving anaphase B spindle elongation to contribute to chromosome segregation (Fig. 4a) [53, 55, 56].
Possible modes of action of motors in the mitotic spindle. a Sliding filament mechanism: motors on anti-parallel overlaps generate force to pull or push the MTs and thereby exert forces on the spindle poles. For example, kinesin-5-dependent outward sliding of ipMTs can push spindle poles apart leading to anaphase B spindle elongation which contributes to chromosome segregation (see Figs. 7b and 8). b Cortical pulling: dynein motors anchored at the actin cortex generate outward forces on the spindle poles, by walking towards the aMTs minus ends anchored at the poles. In some systems, e.g., C. elegans embryos, anaphase B depends mainly on such cortical force generators pulling the poles outward. c Transport motors, e.g., kinetochore dynein transporting kinetochore spindle assembly checkpoint proteins (e.g., Mad2) away from the kinetochore to mediate the transition from metaphase to anaphase. Kinetochore dynein-driven kinetochore motility may also contribute directly to anaphase A in some systems (see Figs. 6 and 7a). d MT-depolymerases, e.g., a member of the kinesin-13 family is shown at the spindle pole depolymerizing the minus ends of MTs as they slide into the poles, to maintain the metaphase spindle length, and contributing to the poleward flux of MTs. In some systems, this process can control the timing and rate of anaphase B (see Figs. 7b and 8). Kinesin-13 depolymerases can also contribute to pacman and poleward flux mechanisms of anaphase A (see Fig. 6). e Sliding against a stable structure: sliding motors (kinesin-5) attached to a stable structure, ‘the spindle matrix’, slide MTs against this fixed structure to generate outward forces on the spindle poles and chromosomes. f MT-binding preference of kinesin-5: the kinesin-5, KLP61F, has a threefold higher binding affinity for anti-parallel MT overlaps compared with parallel MT overlaps
Genetic approaches
In parallel with these targeted biochemical studies, several mitotic motors were uncovered via the analysis of mutants that turned out to encode mitotic kinesins. For example, genes encoding essential kinesin-5 motors were identified in organisms including Aspergillus nidulans [57], budding yeast [58], Xenopus [59] and Drosophila [60]. Loss-of-function mutants were characterized by collapsed monoastral spindles, supporting the hypothesis that kinesin-5 could drive pole–pole separation and spindle elongation. More recently, system-level, genome-wide analyses led to the identification of the full inventory of MT-based motors, including mitotic motors, in several organisms (e.g., [61–63]). For example, in Drosophila melanogaster, there are 36 MT-based motors, 12 of which belong to the dynein family and 24 are members of the kinesin family. Of these, only one dynein (cytoplasmic dynein) and 10–12 kinesins have mitotic functions.
Mitotic motors function by a variety of mechanisms [64, 65]. Below, we arbitrarily classify them into MT crosslinking/sliding, kinetochore, MT depolymerizing and chromosome arm motors, even though mitotic motors often combine these characteristics (e.g., some kinesin-13 motors such as KLP59C are both kinetochore and MT depolymerizing motors).
MT-crosslinking and sliding motors
Kinesin-5
The kinesin-5 family of mitotic motors has been the focus of many studies due to the essential mitotic functions of kinesin-5 in many systems. Kinesin-5 motors are generally thought to be “slow” bipolar homotetramers capable of crosslinking and sliding adjacent spindle MTs (Fig. 4a) [53, 55, 59, 66]. Kinesin-5 could function by sliding MTs against a static structure, for example, a spindle matrix [67], or it could crosslink adjacent MTs and slide anti-parallel MTs apart to generate outward forces on spindle poles to drive anaphase B spindle elongation [55]. While in vivo evidence supports a role for kinesin-5 in anaphase B in some systems [68–70], there appear to be exceptions, since in C. elegans embryos, cortical pulling drives anaphase B [71, 72], whereas in S. pombe, the sliding apart of AP ipMTs by kinesin-6 motors is proposed to drive anaphase B [73].
Recently, using in vitro motility assays, both Xenopus Eg5 and Drosophila KLP61F were shown to crosslink and slide MTs in relation to one another, thus possessing biochemical properties consistent with the latter sliding filament model [74, 75]. In their pioneering studies, Kapitein et al. [74] used a “polymer brush” to force kinesin-5 to bind to immobilized MTs on a glass coverslip instead of to the coverslip surface itself. When the motor crosslinked the immobilized MT to a second, cargo MT in a parallel orientation, the motor apparently moved along both MTs simultaneously without producing any MT–MT sliding, but if the second MT was bound in an antiparallel orientation instead, MT–MT sliding occurred at twice the rate that MTs are moved by kinesin-5 over a glass surface in conventional MT gliding assays [59, 74], strongly supporting the notion of a bipolar motor-driven sliding filament mechanism. In [75], using polarity marked MTs, it was further shown that KLP61F has a distinct MT orientation preference, displaying a threefold preference for bundling MTs in the anti-parallel versus the parallel orientation (Fig. 4f). These findings suggest that in the mitotic spindle, kinesin-5 should accumulate on anti-parallel MT overlaps that are present at the spindle midzone, where it could crosslink and slide apart anti-parallel MTs to generate outward forces on the spindle poles during anaphase B.
To study kinesin-5 dynamics, transgenic flies stably expressing functional GFP-KLP61F, were generated [76]. FRAP and FSM analysis of these lines revealed that kinesin-5 motors exhibit the same turnover dynamics as tubulin in the spindle (with a half-life of ~5 s), and that even though the run-length of the motors on the MTs is short, most motors are bound (85%) to spindle MTs with only a small fraction diffusing freely (15%) in the spindle at any given time. Furthermore, quantitative analysis of the tracks of small groups of motors on MTs (i.e. speckles of motors) indicated that, while tracks of motors on the two half spindles move towards and away from the spindle equator, a large fraction of the motors’ tracks in the central spindle remain stationary. Similar observations were made in Xenopus spindles [77]. This is consistent with the model that, in the central region of the spindle, motors which are bound to anti-parallel MT overlaps slide them apart, while they remain stationary with respect to a fixed laboratory reference frame. In this way, kinesin-5 motors could function as ensembles of dynamic MT–MT crosslinkers that drive anaphase B spindle elongation, thereby contributing to chromosome segregation.
Kinesin-14
The kinesin-14s are dimeric motors (Ncd in Drosophila, Kar3 in budding yeast and Pkl1 in fission yeast) possessing a nucleotide-insensitive MT binding site on their tail domain, and are unique among the kinesin family in that they move towards a MT’s minus end [78–80]. TIRF microscopy motility assays of single GFP-Ncd molecules in buffers approaching physiological ionic strength revealed low processivity along single MTs but enhanced processivity along MT bundles; within the bundles, the K-rich tail of Ncd is proposed to be able to form ionic bonds to “E-hooks” on the cargo MT which reduces Ncd’s dissociation from the MTs, thereby increasing its processivity [81]. These findings support a role for kinesin-14 in bundling and sliding MTs against one another, and generating antagonistic forces relative to kinesin-5 (Fig. 4a). More recently, in careful MT-MT sliding assays, Ncd was shown to crosslink parallel MTs and to crosslink and slide anti-parallel MTs in relation to one another [82]. These assays support a role for kinesin-14 motors both in focusing the spindle poles [83] and in causing spindle collapse in the absence of the antagonistic kinesin-5 motors [69, 84]. In relation to chromosome movements, in Drosophila embryos, it is thought that the kinesin-14, Ncd, exerts a “braking” effect on spindle pole separation during early mitosis, and this gets downregulated at the onset of anaphase B, allowing kinesin-5 motors to drive spindle elongation [26] although this idea is still not universally accepted [85].
Kinetochore motors
A complex set of motors residing at the kinetochore is responsible for (1) the removal of proteins from the MT-attached kinetochores to silence the spindle assembly checkpoint (SAC) and allow the passage from metaphase to anaphase, and/or (2) in powering the polewards or anti-polewards motion of the kinetochores (Fig. 4c). Notable among these proteins is dynein, which moves rapidly (at ~1 μm s−1) towards the MT minus ends. Dynein depletion causes multiple defects during mitosis, including defects in spindle assembly and chromosome congression, in mitotic progression, and in chromosome segregation [86–91]. While a role for dynein in poleward kinetochore transport during ‘search-and-capture’ is generally accepted [7, 92], a direct role for dynein in driving poleward kinetochore transport during anaphase A has been controversial. However, in spindles that utilize a “pacman” mechanism in which kinetochores use kinesin-13 MT depolymerases to “chew” their way to the poles, dynein is thought to help insert the plus ends of kMTs into the kinetochore where the depolymerases reside [86, 93].
The kinesin-7, Cenp-E, is a plus end-directed kinetochore motor which moves slowly and processively along MTs (speed ≈0.01 μm/s, run-length ≈1–5 μm) and is essential for mitosis [94, 95]. It is proposed to maintain kinetochore attachment to depolymerizing/polymerizing kMT plus ends, and to recruit, activate or inactivate cell cycle regulatory proteins to the kinetochores [96, 97]. It can also directly move the kinetochores of unattached or mono-oriented chromosomes along spindle MTs (kMTs or ipMTs) towards the spindle equator to mediate their congression at the metaphase plate [98, 99]. Based on its 230-nm-long, flexible stalk, visible by rotary shadow EM of purified Cenp-E [95], it has been proposed that Cenp-E could act as a flexible tether to MT plus ends, thus forming the “kinetochore fibrils” that are seen in vivo at the MT-kinetochore interface in high resolution EM tomography studies [100], although the Ndc-80 protein complex is an alternative candidate [101–106]. The linkage of kinetochores to depolymerizing MT-plus ends via a highly flexible, plus end-directed motor with load-bearing capacity, such as Cenp-E, is highly appealing [95, 96, 100, 107].
MT-depolymerizing motors
Kinesin-13 motors
The kinesin-13 family of motors bind to the MT ends, where they use the free energy released by ATP hydrolysis to remove tubulin subunits and induce MT depolymerization (Fig. 4d) [108, 109]. In vivo, the depletion of the vertebrate kinesin-13, MCAK, leads to chromosome congression and segregation defects [110, 111], and in elegant studies, the Sharp laboratory have shown that two Drosophila kinesin-13 motors cooperate to drive rapid (0.1 μm s−1), poleward chromosome motility by depolymerizing opposite ends of the kMTs, producing a combined “flux-pacman” mechanism for anaphase A in the syncytial embryo [93]. Recent in vitro studies have illuminated the mechanism of action of kinesin-13 depolymerases. For example, the vertebrate kinesin-13, MCAK, was shown to bind to the MT lattice wall and to use thermal energy to diffuse to the ends of the MTs via a 1D random walk independent of ATP; only when it reaches a MT’s tip does it use ATP hydrolysis to catalyze the removal of one or more tubulin dimers from the tip [112]. These motors appear to either increase or decrease the frequency of MT plus end catastrophe or rescue, respectively and the resulting deploymerase activity is regulated by phosphorylation [113–115]. At least one kinesin-13, KLP10A, forms rings around the MT lattice that are visible by EM, and a model based on high resolution structural analysis suggests that the MT binding loop 2 of the conserved kinesin superfamily motor domain contains kinesin-13-specific residues which stabilize MT protofilaments in the “bent” conformation, characteristic of depolymerization [116, 117]. Thus, kinesin-13 motors, like the Dam/Dash complex, are candidates for serving as coupling rings between depolymerizing MT ends and the kinetochore or the spindle pole [116].
Kinesin-8 motors
Kinesin-8 motors are of great interest since they use energy from ATP hydrolysis to translocate along the MT polymer lattice towards the MT plus ends, and also, when they arrive at the tip, to induce MT-depolymerization. The depletion of kinesin-8 motors from dividing cells leads to mitotic defects that include aberrant congression and directional instability of chromosomes and the formation of abnormally long spindles [118–124]. Recent in vitro studies reveal that the yeast kinesin-8, Kip3p, lands on the MT-lattice wall then translocates to the MT-plus end where it pauses until another kinesin-8 arrives and bumps it off the MT tip, allowing it to peel off one or two tubulin dimers as it dissociates [125]. This sequence of events renders the kinesin-8 motor a “length-dependent-depolymerase” because relatively few kinein-8 motors will land on short MTs so that only a few motors will accumulate at their plus ends to depolymerize them, whereas a larger number of these motors will land on longer MTs and accumulate at their plus ends, leading to more frequent depolymerization events. This mechanism may be crucial in controlling the distribution of MT lengths in the spindle by modulating the catastrophe frequency and/or the depolymerization rate of MT plus ends. Thus, it will be interesting to determine the influence of kinesin-8 on the MT length distribution and average MT length 〈L〉, which is given by 〈L〉 = (V s V g)/(V s f cat − V g f res) when V s f cat > V g f res and for fixed rates V s , V g, f cat and f res [126, 127].
Motors linking the chromosome arms to spindle MTs
A variety of kinesin motors are bound to the arms of mitotic chromosomes where they translocate chromosomes along spindle MTs to contribute to spindle assembly and chromosome positioning [128, 129]. For example, in Drosophila, the kinesin-10, Nod, uses a DNA-binding domain in its tail to bind to chromosome arms where its motor domains enhance spindle MT plus end polymerization, allowing the growing MTs to push chromosomes to the spindle equator by a polymer ratchet mechanism [130]. Indeed, the ATPase cycle of this motor is such that it can potentially mediate a perfect ‘clamped-filament elongation’ mechanism [131]. Based on elegant structural and biochemical studies, a model has been proposed for the mechanism by which the ATP hydrolysis cycle of ensembles of chromosome-associated Nod proteins can maintain the attachment of a chromosome to the growing MT plus ends, thereby contributing to the “polar ejection force” which pushes chromosomes to the equator during chromosome capture and congression [130].
Cooperation between mitotic force generators
Mitotic motors seldom work in isolation, but instead, ensembles of multiple mitotic motors and dynamic MT polymer ratchets cooperate in subtle and sophisticated ways to generate the balance of forces that coordinate chromosome movements in the spindle [64, 65].
Antagonism and cooperation between opposite polarity MT sliding motors
In one, classic example of motor cooperation, plus end and minus end-directed MT-based motors crosslink and slide adjacent ipMTs inward and outward to maintain the position of the spindle poles. For example, kinesin-5 and kinesin-14 motors bound to the anti-parallel overlap region of MTs antagonize one another to exert outward and inward forces on spindle poles that can assemble or maintain the prometaphase spindle in some systems (Fig. 5a) [58, 66, 69]. In a variant of this mechanism, it has been proposed that dynein substitutes for kinesin-14 in some spindles [132]. An alternative mechanism by which plus and minus end-directed motors (e.g., kinesin-5 and dynein) can cooperate is the so-called “slide-and-cluster” mechanism in which kinesin-5 motors slide anti-parallel MTs apart whereas dynein clusters the MTs’ non-dynamic minus ends—and opposes sliding—once the sliding MTs’ minus ends reach the spindle poles [28] (Fig. 5b). Opposite polarity MT-based motors (e.g., kinesin-5 and dynein) can also cooperate or antagonize one another by acting on distinct sets of spindle MTs. For example, cortical dynein can pull on astral MTs, thereby pulling apart spindle poles, and this activity is thought to augment the kinesin-5-driven sliding apart of ipMTs which pushes apart the poles during late anaphase B in the Drosophila embryo [69] (Fig. 5d). In contrast, in the early C. elegans embryo, it is thought that spindle elongation due to rapid dynein-generated pulling forces on aMTs (Fig. 4b) is restrained by the braking effect of kinesin-5 on ipMTs [71, 72] (Fig. 5e).
Functional coordination of mitotic motors. a Sliding motors can antagonize one another to maintain pole–pole spacing (S(t)) when dS/dt = 0, elongate the spindle (dS/dt > 0) or shorten the spindle (dS/dt < 0). The outcome depends on the relative ratio of active and bound motors on the anti-parallel overlaps. b Slide and cluster model: dynein homo-dimers and kinesin-5 cooperate near the chromosomes where MTs mainly overlap anti-parallel (both motors slide MTs apart) and antagonize one another near the poles where MTs overlap mainly parallel (dynein clustering the minus ends opposes sliding by kinesin-5). This model can explain the control of anastral mitotic spindle length and poleward flux of MTs in the absence of MT minus end depolymerization activity. c Depolymerizing (e.g., kinesin-13 on spindle poles) and sliding motors (e.g., kinesin-5 and kinesin-14 on AP ipMT overlaps) can cooperate to maintain spindle length during metaphase, and give rise to poleward flux of MTs (see Figs. 7b and 8). d Kinesin-5 acting on anti-parallel overlaps and cortical dynein acting on aMTs can cooperate during anaphase B to elongate the spindle. For example, in early anaphase B in the Drosophila embryo, the anti-parallel overlaps are abundant and kinesin-5-generated forces drive spindle elongation. In late anaphase B, the anti-parallel overlap is diminished as the spindle length increases, and more aMT plus ends can reach the cortex, whereupon dynein-generated pulling forces assist the kinesin-5 driven outward forces to complete anaphase B chromosome segregation. e Kinesin-5 on anti-parallel ipMT overlaps and cortical dynein on aMTs can compete during anaphase B spindle elongation in some organisms. For example, in C. elegans embryos, the cortical dynein-driven rapid pulling apart of spindle poles is resisted by the slower kinesin-5 motors bound to the ipMT overlaps which may act as ‘brakes’ to slow down the rate of anaphase B spindle elongation
Functional cooperation between MT-motors and MT polymer dynamics
The outward sliding apart of ipMTs (e.g., due to kinesin-5) can be antagonized by kinesin-13 MT-depolymerases located at the spindle poles where they use ATP hydrolysis to remove tubulin subunits from the minus ends of the MTs as they are being slid polewards. This can result in the production of poleward flux and the maintenance of a constant or steady state spindle length (Fig. 5c) [26, 68, 93]. In addition, forces due to MT polymerization or depolymerization, or elastic forces due to the stretching or buckling of spindle MTs, can pull and push the chromosomes and spindle poles. For example, it has been proposed that the growing plus ends of MTs that extend from one spindle pole can impinge on the opposite pole where they produce outward-directed polymerization ratcheting forces, which cooperate with kinesin-14 and cortical dynein-dependent forces during early spindle assembly [133]. In addition, as described above, the chromosomal arm kinesin-10, Nod, cooperates with MT polymerization to generate the polar ejection forces that exert plus end-directed pushing forces on chromosome arms [130].
Mechanism of metaphase and anaphase A chromosome dynamics
Where and how the force driving poleward chromosome motility is generated has been a topic of research and vigorous debate since Flemming’s early observations of mitosis [1], and several models have been proposed, including those discussed below. Several very early models were discussed by Schrader [134].
1. Forces are generated all along the kinetochore fiber (Fig. 6a)
(a) In the ‘Traction Fiber’ model, the number of force generators per unit length is constant and consequently the longer the kMT, the higher is the force pulling a chromosome poleward [135]; and (b) a ‘Sliding Filament Model’ was proposed in which the sliding of adjacent spindle MTs mediates anaphase A, although the aforementioned EM analysis of the three-dimensional fine structure of the spindle by McIntosh et al. [50] explicitly eliminated this model as applied to chromosome-to-pole movement.
Models for anaphase A chromosome-to-pole motility. a The traction fiber model: longer kMTs can generate higher poleward forces on the chromosome. b Forces due to MT dynamics. Top panel in the dynamic equilibrium model, during anaphase, kMTs undergo depolymerization to pull chromosomes to poles (anaphase A) whereas ipMTs polymerize to push apart the spindle poles (anaphase B). Lower panel the Hill–Sleeve model: high binding affinity of MTs in the kinetochore sleeve mediates either pushing or pulling of the chromosome to attain the minimal energy conformation (i.e. maximum number of weak bonds) of sleeve-MT interactions during MT polymerization and depolymerization, respectively. c Pulling forces at the pole: a force-generator located at the pole is generating force to reel in the kMT and pull the chromosome polewards. Here, the MT minus end may be depolymerized at the pole or may slide past the pole. d The combined flux–pacman mechanism: the kMT depolymerizes both at its kinetochore attached plus and pole attached minus end, while the kMT fluxes polewards. e The slip–clutch mechanism at the kinetochore. The kMT plus end switches to polymerization when a threshold (high) tension between the sister kinetochores is reached, and reverses to depolymerization when the tension is reduced below a threshold (low). f Uniform tension among MTs and sister kinetochores are attained by persistent poleward fluxing of interconnected spindle MTs. In the absence of poleward flux, the tension variance is maintained over long time scales
2. Forces are generated by kMT depolymerization (Fig. 6b)
(i) In the ‘Dynamic Equilibrium Model’, spindle MTs polymerize when the spindle is forming and elongating but they transition to depolymerization to drive chromosome-to-pole movement [34]; (ii) in the ‘Treadmilling Model’ proposed by Margolis and Wilson, the kMT shortens by slow (or no) addition of tubulin dimers at the plus ends combined with their removal at the minus ends [136]; subsequently the ‘Dynamic equilibrium model’ was revised by Inoué to accommodate the depolymerization of MT minus ends at the poles [137]; and (iii) in the ‘Hill sleeve’ model, depolymerizing kMT plus ends maintain attachment with the kinetochore, consistent with thermodynamics [138]. This model was further explored in the context of a force–balance mechanism, and was shown to account strikingly well for the oscillations of bi- and mono-oriented chromosomes in PtK cells during metaphase and prometaphase [139].
3. Forces are generated at the spindle poles (Fig. 6c)
Based on observations of kMT poleward flux and tension-dependent sister kinetochore distance in taxol treated newt lung cell spindles, Salmon et al. [140] suggested the presence of kinetochore pulling forces generated at the spindle poles.
4. The combined “flux–pacman” mechanism (Fig. 6d)
For example, the functional perturbation of two distinct kinesin-13 MT-depolymerases located at the spindle pole (KLP10A) and at the kinetochore (KLP59C) supported a combined flux–pacman mechanism of anaphase A [93]. Dynein located at the kinetochore is thought to augment this mechanism by inserting the plus ends of the kMTs into the jaws of the kinetochore-bound kinesin-13 (KLP59C) to facilitate plus end depolymerization. In this mechanism, the depolymerization of MT minus ends by a kinesin-13, KLP10A, located at the spindle pole is thought to be coupled either to a force generator at the poles that reels in the kMTs or to the kinesin-5-driven poleward sliding of ipMTs which may be crosslinked to passively sliding kMTs [68, 141]. Recently, a third kinesin-13 and several MT severing proteins have been proposed to augment this basic mechanism [142, 143].
5. Tension-based regulation of metaphase chromosome positioning (Fig. 6e)
Salmon et al. have proposed a molecular mechanism for the regulation of kMT plus end dynamics by tension forces generated between sister kinetochores [31, 144]. In this mechanism, high tension promotes polymerization of the kMT plus end, while low tension promotes depolymerization. In an elegant computational approach, a model accounting for this mechanism successfully explained the congression of the chromosomes in budding yeast mitosis [145]. Furthermore, this model made an important prediction, namely the presence of a spatial gradient of MT plus end catastrophe events, with high catastrophe rates at the spindle equator. Subsequently, the authors have proposed that the kinesin-5, Cin8, is involved in the establishment of this catastrophe gradient, but how it does so is unknown [146].
6. Synchrony of chromosomes during anaphase A (Fig. 6f)
In a recent experimental and theoretical study, the synchrony of chromatid-to-pole movement during anaphase A was proposed to depend upon the poleward flux of spindle MTs (both ipMT and kMTs), resulting from their kinesin-5 dependent outward sliding coupled to kinesin-13 dependent minus end depolymerization with elastic couplers linking the spindle MTs to one another [147]. The authors argue that the poleward sliding of spindle MTs during metaphase leading to an evolving uniform extension/compression of the linkers along the spindle axis and transmitted to all kMTs in the spindle orchestrates a uniform tension at the kinetochores which underlies the synchronous entry and progress of chromosome-to-pole movement during anaphase A.
Based on the molecular models discussed above, several quantitative models have been proposed that are aimed at improving our understanding of the mechanisms underlying chromosome dynamics at various stages of mitosis, from metaphase through anaphase A, and a scholarly review of these models can be found in [148]. For example, a quantitative force–balance model based on the above “flux–pacman” mechanism for poleward chromosome motility was developed for fly embryo mitosis, Fig. 7a [149], which proposes that: (1) the same kinetochore machinery and underlying mechanism coordinates the dynamic behavior of chromosomes during metaphase and anaphase A, so that the transition from metaphase to anaphase A occurs simply through the degradation of the cohesin bonds between sister kinetochores and the removal of PE forces; and (2) differences in the dynamics of chromosomes in different organisms, including the amplitude and frequency of their oscillations and rates of their movement as well as differences in the relative contribution of the flux and pacman components of their pole-directed motility, represent adaptations of the same underlying mechanism.
Quantitative models for anaphase A and B in the Drosophila Embryo. Both models depend on a balance of forces in which the net force (F net(t)) acting on a chromosome or spindle pole moves it at a velocity v = v(t) against viscous drag (μ or ζ) under low Reynolds number conditions, where F net = μv or ζv. a The movement and positioning of chromosomes during metaphase and anaphase A is determined by a balance of forces that is exerted on them by various motors acting on MTs, kinetochores, chromosome arms, or spindle poles. MT dynamics, tension due to cohesins linking sister chromatids and the motors’ force–velocity curves are also accounted for in the model (see [149] for details). b Spindle pole dynamics during pre-anaphase B (i.e. the metaphase/anaphase A steady state spindle length) and anaphase B (when the spindle elongates) is described by a system of equations based on three core equations. These include two kinematic equations which describe changes in spindle length (S(t)) and ipMT overlap length (L(t)) over time, and a force–balance equation which describes the rate of pole movement resulting from the cumulative effect (assumed to be additive) of many motors that act on ipMT overlaps, obeying linear force–velocity curves. The model incorporates realistic spindle dynamics and geometry as well as a pole-associated MT depolymerase acting as a switch to turn on spindle elongation (Fig. 8) (see [26] for details). Both models represent “working models” that we find useful for identifying key components or parameters to be measured, and for designing better experiments to test the validity of our current ideas, which at best only approximate the actual molecular mechanisms underlying this complex and fascinating process
Mechanism of anaphase B
Based on experimental evidence from many organisms ranging from budding yeast to vertebrates, it is now well-established that a sliding filament mechanism similar to that proposed by McIntosh in 1969 underlies spindle elongation during anaphase B [14, 26, 68–70, 150–153]. Recent work has led to a model in which kinesin-5 motors drive the sliding apart of anti-parallel ipMTs to exert force on the spindle poles, whereas poleward flux serves as an on–off switch (Fig. 8).
Model for anaphase B. a The transition from poleward flux during pre-anaphase B (i.e. metaphase/anaphase A) to spindle elongation at the onset of anaphase B. The cartoon is an unrealistic simplification in which only a single pair of overlapping AP ipMTs is shown. b The switch in the dynamics of MT plus and minus ends mediated by cyclin B degradation is essential for steady, linear and robust anaphase B chromosome segregation. This involves: (1) the down-regulation of ipMT minus end depolymerization at spindle poles which turns off poleward flux, allowing outwardly sliding ipMTs to exert pushing forces on the poles; and (2) the onset of a gradient of MT catastrophe frequencies (f cat) which causes ipMT plus ends (marked by + tip trackers like EB1) to grow and invade the spindle equator, thereby building a robust midzone comprising multiple overlapping AP ipMT plus ends which are slid apart at a linear rate by kinesin-5 motors to accomplish complete spindle elongation
This model is based on FSM analysis of Drosophila embryo mitotic spindles which revealed that spindle MTs persistently slide apart, so that tubulin speckles move away from the equator, throughout metaphase and anaphase. However, the pole–pole distance is maintained constant, giving rise to poleward flux of fluorescent tubulin speckles prior to anaphase B, but, following the onset of anaphase B, the poles move away from the equator at the same rate as the speckles (Fig. 2c and 8a) [30]. This evidence suggested that: (1) the motor responsible for anaphase B must also be responsible for the sliding of ipMTs driving polewards flux; and (2) it is a change in the dynamics of the minus ends of the MTs which leads to the switch from the steady positioning of the poles to spindle elongation. In this system, the kinesin-5, KLP61F, is thought to crosslink and slide apart anti-parallel ipMT throughout metaphase, anaphase A and anaphase B spindle elongation [68, 69, 154], while the kinesin-13, KLP10A, is responsible for the depolymerization of the MT minus ends at the spindle poles prior to anaphase B [93]. Furthermore, ipMTs in the equatorial region of the spindle at anaphase B were extremely dynamic, turning over with a half-life of ~5 s, and the inhibition of the kinesin-4, KLP3A, revealed an inverse linear correlation between poleward flux and spindle elongation rates during anaphase B, indicating that the extent of the decrease in the rate of ipMT depolymerization at the poles can regulate the rate, as well as the extent, of anaphase B spindle elongation [26, 152].
A mathematical model (Fig. 7b) was developed to address many intriguing aspects of the anaphase B molecular mechanism. For example, how could the bipolar motors working on highly dynamic MTs maintain the observed linear rate of spindle elongation, and what biophysical and kinetic properties of the molecules involved in this mechanism affect the anaphase B rate [26]? The quantitative model revealed that: (1) motors working on highly dynamic anti-parallel MT overlaps can drive a steady-linear elongation of the spindle; (2) the bipolar motors are working near their ‘unloaded regime’ during this process; (3) the rate of the spindle elongation is determined only by the unloaded sliding rate of the bipolor motors and the extent of suppression of poleward flux; (4) a sustained MT plus end polymerization is necessary to sustain robust elongation of the spindle. Several of these predictions were subsequently tested experimentally [68, 152]. For example, it was determined that cyclin B degradation leads to the suppression of poleward flux, and also induces an ipMT “catastrophe” gradient which causes ipMTs to undergo net polymerization to build a robust overlap zone to sustain robust linear spindle elongation, as predicted [152]. Our current “working model” for the mechanism of anaphase B spindle elongation is shown in Fig. 8b.
It is clear that the mechanism presented in Fig. 8b is not used ubiquitously. For example, in the C. elegans embryo, the kinesin-5, BMK-1, is not essential. Here, cortical dynein motors pull on astral MTs to pull apart the spindle poles and drive anaphase B spindle elongation, while kinesin-5 acts on the AP ipMT overlaps to serve as a brake that controls the rate of spindle elongation (Figs. 4b and 5e) [72]. Therefore, even though in some organisms a sliding filament mechanism driven by bipolar kinesin-5 motors underlies anaphase B spindle elongation, natural selection has produced alternative mechanisms in different systems, as appears to be the case for many aspects of mitosis.
Conclusion
Work done during the many decades that have elapsed since Flemming’s description of mitosis has led to the identification of several key force-generating mechanisms that cooperate to ensure accurate chromosome movement and segregation during mitosis. Although satisfying progress has been made, our understanding of the mechanism of force generation for chromosome movements remains incomplete and much remains to be learned. Thus, mitosis enthusiasts can expect to remain active for many decades into the future.
References
Flemming W (1882) Zellsubstanz, Kern und Zelltheilung, Vogel. Leipzig
Nicklas RB (1988) The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem 17:431–449
Dumont S, Mitchison TJ (2009) Force and length in the mitotic spindle. Curr Biol 19:R749–R761
Berg HC (1983) Random walks in biology. Princeton University Press, Princeton
Howard J (2001) Mechanics of motor proteins and the cytoskeleton. Sinauer, Sunderland
Marshall WF, Marko JF, Agard DA, Sedat JW (2001) Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr Biol 11:569–578
Alexander SP, Rieder CL (1991) Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J Cell Biol 113:805–815
Nicklas RB (1983) Measurements of the force produced by the mitotic spindle in anaphase. J Cell Biol 97:542–548
Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW (1997) Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7:930–939
Ke K, Cheng J, Hunt AJ (2009) The distribution of polar ejection forces determines the amplitude of chromosome directional instability. Curr Biol 19:807–815
Amos L, Klug A (1974) Arrangement of subunits in flagellar microtubules. J Cell Sci 14:523–549
Crepeau RH, McEwen B, Dykes G, Edelstein SJ (1977) Structural studies on porcine brain tubulin in extended sheets. J Mol Biol 116:301–315
Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96:79–88
McDonald KL, Edwards MK, McIntosh JR (1979) Cross-sectional structure of the central mitotic spindle of Diatoma vulgare. Evidence for specific interactions between antiparallel microtubules. J Cell Biol 83:443–461
McDonald KL, O’Toole ET, Mastronarde DN, McIntosh JR (1992) Kinetochore microtubules in PTK cells. J Cell Biol 118:369–383
McIntosh JR, Roos UP, Neighbors B, McDonald KL (1985) Architecture of the microtubule component of mitotic spindles from Dictyostelium discoideum. J Cell Sci 75:93–129
Ding R, McDonald KL, McIntosh JR (1993) Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol 120:141–151
Euteneuer U, McIntosh JR (1981) Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol 89:338–345
Goshima G, Kimura A (2009) New look inside the spindle: microtubule-dependent microtubule generation within the spindle. Curr Opin Cell Biol. doi:10.1016/j.ceb.2009.11.012
Inoue S (1953) Polarization optical studies of the mitotic spindle. I. The demonstration of spindle fibers in living cells. Chromosoma 5:487–500
Waterman-Storer C, Desai A, Salmon ED (1999) Fluorescent speckle microscopy of spindle microtubule assembly and motility in living cells. Methods Cell Biol 61:155–173
Saxton WM, Stemple DL, Leslie RJ, Salmon ED, Zavortink M, McIntosh JR (1984) Tubulin dynamics in cultured mammalian cells. J Cell Biol 99:2175–2186
Mitchison TJ (1989) Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J Cell Biol 109:637–652
Zhai Y, Kronebusch PJ, Borisy GG (1995) Kinetochore microtubule dynamics and the metaphase–anaphase transition. J Cell Biol 131:721–734
Cassimeris L, Inoue S, Salmon ED (1988) Microtubule dynamics in the chromosomal spindle fiber: analysis by fluorescence and high-resolution polarization microscopy. Cell Motil Cytoskeleton 10:185–196
Brust-Mascher I, Civelekoglu-Scholey G, Kwon M, Mogilner A, Scholey JM (2004) Model for anaphase B: role of three mitotic motors in a switch from poleward flux to spindle elongation. Proc Natl Acad Sci USA 101:15938–15943
Maiato H, Khodjakov A, Rieder CL (2005) Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol 7:42–47
Burbank KS, Mitchison TJ, Fisher DS (2007) Slide-and-cluster models for spindle assembly. Curr Biol 17:1373–1383
Maddox P, Desai A, Oegema K, Mitchison TJ, Salmon ED (2002) Poleward microtubule flux is a major component of spindle dynamics and anaphase a in mitotic Drosophila embryos. Curr Biol 12:1670–1674
Brust-Mascher I, Scholey JM (2002) Microtubule flux and sliding in mitotic spindles of Drosophila embryos. Mol Biol Cell 13:3967–3975
Maddox P, Straight A, Coughlin P, Mitchison TJ, Salmon ED (2003) Direct observation of microtubule dynamics at kinetochores in Xenopus extract spindles: implications for spindle mechanics. J Cell Biol 162:377–382
LaFountain JR Jr, Cohan CS, Siegel AJ, LaFountain DJ (2004) Direct visualization of microtubule flux during metaphase and anaphase in crane-fly spermatocytes. Mol Biol Cell 15:5724–5732
Mitchison TJ (2005) Mechanism and function of poleward flux in Xenopus extract meiotic spindles. Philos Trans R Soc Lond B 360:623–629
Inoue S, Sato H (1967) Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J Gen Physiol 50(suppl):259–292
Borisy GG, Taylor EW (1967) The mechanism of action of colchicine. Binding of colchincine-3H to cellular protein. J Cell Biol 34:525–533
Borisy GG, Taylor EW (1967) The mechanism of action of colchicine. Colchicine binding to sea urchin eggs and the mitotic apparatus. J Cell Biol 34:535–548
Weisenberg RC (1972) Microtubule formation in vitro in solutions containing low calcium concentrations. Science 177:1104–1105
Mogilner A, Oster G (2003) Polymer motors: pushing out the front and pulling up the back. Curr Biol 13:R721–R733
Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312:237–242
Dogterom M, Yurke B (1997) Measurement of the force–velocity relation for growing microtubules. Science 278:856–860
Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR (2005) Force production by disassembling microtubules. Nature 438:384–388
Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H (2006) Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci USA 103:5741–5745
Valentine MT, Fordyce PM, Krzysiak TC, Gilbert SP, Block SM (2006) Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat Cell Biol 8:470–476
Gibbons IR, Rowe AJ (1965) Dynein: a protein with adenosine triphosphatase activity from cilia. Science 149:424–426
Vale RD, Reese TS, Sheetz MP (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 42:39–50
Lasek RJ, Brady ST (1985) Attachment of transported vesicles to microtubules in axoplasm is facilitated by AMP-PNP. Nature 316:645–647
Lye RJ, Porter ME, Scholey JM, McIntosh JR (1987) Identification of a microtubule-based cytoplasmic motor in the nematode C. elegans. Cell 51:309–318
Paschal BM, Shpetner HS, Vallee RB (1987) MAP 1C is a microtubule-activated ATPase which translocates microtubules in vitro and has dynein-like properties. J Cell Biol 105:1273–1282
Paschal BM, Vallee RB (1987) Retrograde transport by the microtubule-associated protein MAP 1C. Nature 330:181–183
McIntosh JR, Hepler PK, Van Wie DG (1969) Model for mitosis. Nature 224:659–663
Scholey JM, Porter ME, Grissom PM, McIntosh JR (1985) Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature 318:483–486
Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM (1992) Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J Cell Sci 101:291–301
Cole DG, Saxton WM, Sheehan KB, Scholey JM (1994) A “slow” homotetrameric kinesin-related motor protein purified from Drosophila embryos. J Biol Chem 269:22913–22916
Wright BD, Terasaki M, Scholey JM (1993) Roles of kinesin and kinesin-like proteins in sea urchin embryonic cell division: evaluation using antibody microinjection. J Cell Biol 123:681–689
Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM (1996) A bipolar kinesin. Nature 379:270–272
Kashina AS, Scholey JM, Leszyk JD, Saxton WM (1996) An essential bipolar mitotic motor. Nature 384:225
Enos AP, Morris NR (1990) Mutation of a gene that encodes a kinesin-like protein blocks nuclear division in Anidulans. Cell 60:1019–1027
Saunders WS, Hoyt MA (1992) Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70:451–458
Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359:540–543
Heck MM, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LS (1993) The kinesin-like protein KLP61F is essential for mitosis in Drosophila. J Cell Biol 123:665–679
Goshima G, Vale RD (2003) The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 162:1003–1016
Miki H, Setou M, Kaneshiro K, Hirokawa N (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA 98:7004–7011
Stewart RJ, Pesavento PA, Woerpel DN, Goldstein LS (1991) Identification and partial characterization of six members of the kinesin superfamily in Drosophila. Proc Natl Acad Sci USA 88:8470–8474
Loughlin R, Riggs B, Heald R (2008) SnapShot: motor proteins in spindle assembly. Cell 134:548–548
Sharp DJ, Rogers GC, Scholey JM (2000) Microtubule motors in mitosis. Nature 407:41–47
Tao L, Mogilner A, Civelekoglu-Scholey G, Wollman R, Evans J, Stahlberg H, Scholey JM (2006) A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol 16:2293–2302
Kapoor TM, Mitchison TJ (2001) Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J Cell Biol 154:1125–1133
Brust-Mascher I, Sommi P, Cheerambathur DK, Scholey JM (2009) Kinesin-5-dependent poleward flux and spindle length control in Drosophila embryo mitosis. Mol Biol Cell 20:1749–1762
Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM (2000) Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell 11:241–253
Straight AF, Sedat JW, Murray AW (1998) Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol 143:687–694
Grill SW, Gonczy P, Stelzer EH, Hyman AA (2001) Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature 409:630–633
Saunders AM, Powers J, Strome S, Saxton WM (2007) Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol 17:R453–R454
Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT (2009) Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell 17:257–267
Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435:114–118
van den Wildenberg SM, Tao L, Kapitein LC, Schmidt CF, Scholey JM, Peterman EJ (2008) The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol 18:1860–1864
Cheerambathur DK, Brust-Mascher I, Civelekoglu-Scholey G, Scholey JM (2008) Dynamic partitioning of mitotic kinesin-5 cross-linkers between microtubule-bound and freely diffusing states. J Cell Biol 182:429–436
Uteng M, Hentrich C, Miura K, Bieling P, Surrey T (2008) Poleward transport of Eg5 by dynein–dynactin in Xenopus laevis egg extract spindles. J Cell Biol 182:715–726
Karabay A, Walker RA (1999) Identification of microtubule binding sites in the Ncd tail domain. Biochemistry 38:1838–1849
McDonald HB, Stewart RJ, Goldstein LS (1990) The kinesin-like ncd protein of Drosophila is a minus end-directed microtubule motor. Cell 63:1159–1165
Walker RA, Salmon ED, Endow SA (1990) The Drosophila claret segregation protein is a minus-end directed motor molecule. Nature 347:780–782
Furuta K, Toyoshima YY (2008) Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr Biol 18:152–157
Fink G, Hajdo L, Skowronek KJ, Reuther C, Kasprzak AA, Diez S (2009) The mitotic kinesin-14 Ncd drives directional microtubule–microtubule sliding. Nat Cell Biol 11:717–723
Goshima G, Nedelec F, Vale RD (2005) Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J Cell Biol 171:229–240
Sharp DJ, Yu KR, Sisson JC, Sullivan W, Scholey JM (1999) Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat Cell Biol 1:51–54
Hallen MA, Liang ZY, Endow SA (2008) Ncd motor binding and transport in the spindle. J Cell Sci 121:3834–3841
Sharp DJ, Rogers GC, Scholey JM (2000) Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat Cell Biol 2:922–930
Yang Z, Tulu US, Wadsworth P, Rieder CL (2007) Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol 17:973–980
Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155:1159–1172
Schmidt DJ, Rose DJ, Saxton WM, Strome S (2005) Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell 16:1200–1212
Varma D, Monzo P, Stehman SA, Vallee RB (2008) Direct role of dynein motor in stable kinetochore–microtubule attachment, orientation, and alignment. J Cell Biol 182:1045–1054
Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177:1005–1015
Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB (1996) Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol 132:617–633
Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ (2004) Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature 427:364–370
Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW (1991) CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J 10:1245–1254
Kim Y, Heuser JE, Waterman CM, Cleveland DW (2008) CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol 181:411–419
Lombillo VA, Nislow C, Yen TJ, Gelfand VI, McIntosh JR (1995) Antibodies to the kinesin motor domain and CENP-E inhibit microtubule depolymerization-dependent motion of chromosomes in vitro. J Cell Biol 128:107–115
Mao Y, Abrieu A, Cleveland DW (2003) Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114:87–98
Kapoor TM, Lampson MA, Hergert P, Cameron L, Cimini D, Salmon ED, McEwen BF, Khodjakov A (2006) Chromosomes can congress to the metaphase plate before biorientation. Science 311:388–391
Cai S, O’Connell CB, Khodjakov A, Walczak CE (2009) Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol 11:832–838
McIntosh JR, Grishchuk EL, Morphew MK, Efremov AK, Zhudenkov K, Volkov VA, Cheeseman IM, Desai A, Mastronarde DN, Ataullakhanov FI (2008) Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell 135:322–333
DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127:969–982
Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A (2005) Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem 280:29088–29095
Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, Salek M, Rappsilber J, Moores CA, Salmon ED, Musacchio A (2008) Implications for kinetochore–microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133:427–439
Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127:983–997
Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL (2009) The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell 136:865–875
Wan X, O’Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, Stukenberg PT, Desai A, Salmon ED (2009) Protein architecture of the human kinetochore microtubule attachment site. Cell 137:672–684
Lombillo VA, Stewart RJ, McIntosh JR (1995) Minus-end-directed motion of kinesin-coated microspheres driven by microtubule depolymerization. Nature 373:161–164
Desai A, Verma S, Mitchison TJ, Walczak CE (1999) Kin I kinesins are microtubule-destabilizing enzymes. Cell 96:69–78
Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J (2003) The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell 11:445–457
Maney T, Hunter AW, Wagenbach M, Wordeman L (1998) Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol 142:787–801
Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE (2004) Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell 15:1146–1159
Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J (2006) The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature 441:115–119
Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ (2005) Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol 7:235–245
Mennella V, Tan DY, Buster DW, Asenjo AB, Rath U, Ma A, Sosa HJ, Sharp DJ (2009) Motor domain phosphorylation and regulation of the Drosophila kinesin 13, KLP10A. J Cell Biol 186:481–490
Moutinho-Pereira S, Debec A, Maiato H (2009) Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell 20:2796–2808
Tan D, Asenjo AB, Mennella V, Sharp DJ, Sosa H (2006) Kinesin-13s form rings around microtubules. J Cell Biol 175:25–31
Tan D, Rice WJ, Sosa H (2008) Structure of the kinesin13-microtubule ring complex. Structure 16:1732–1739
Gandhi R, Bonaccorsi S, Wentworth D, Doxsey S, Gatti M, Pereira A (2004) The Drosophila kinesin-like protein KLP67A is essential for mitotic and male meiotic spindle assembly. Mol Biol Cell 15:121–131
Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM (2005) Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci 118:2671–2682
Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM (2004) Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci 117:3669–3677
Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L (2008) The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell 14:252–262
Cottingham FR, Gheber L, Miller DL, Hoyt MA (1999) Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J Cell Biol 147:335–350
West RR, Malmstrom T, Troxell CL, McIntosh JR (2001) Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell 12:3919–3932
West RR, Malmstrom T, McIntosh JR (2002) Kinesins klp5(+) and klp6(+) are required for normal chromosome movement in mitosis. J Cell Sci 115:931–940
Varga V, Leduc C, Bormuth V, Diez S, Howard J (2009) Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell 138:1174–1183
Verde F, Dogterom M, Stelzer E, Karsenti E, Leibler S (1992) Control of microtubule dynamics and length by cyclin A- and cyclin B-dependent kinases in Xenopus egg extracts. J Cell Biol 118:1097–1108
Chen YD, Hill TL (1987) Theoretical studies on oscillations in microtubule polymerization. Proc Natl Acad Sci USA 84:8419–8423
Brunet S, Vernos I (2001) Chromosome motors on the move. From motion to spindle checkpoint activity. EMBO Rep 2:669–673
Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C (1995) Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell 81:117–127
Cochran JC, Sindelar CV, Mulko NK, Collins KA, Kong SE, Hawley RS, Kull FJ (2009) ATPase cycle of the nonmotile kinesin NOD allows microtubule end tracking and drives chromosome movement. Cell 136:110–122
Dickinson RB, Purich DL (2002) Clamped-filament elongation model for actin-based motors. Biophys J 82:605–617
Ferenz NP, Paul R, Fagerstrom C, Mogilner A, Wadsworth P (2009) Dynein antagonizes eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol 19:1833–1838
Cytrynbaum EN, Scholey JM, Mogilner A (2003) A force balance model of early spindle pole separation in Drosophila embryos. Biophys J 84:757–769
SchraderF (1944) Mitosis, the movement of chromosomes in cell division. Columbia University Press, New York
Östergren SG (1951) The mechanism of co-orientation in bivalents and multivalents: the theory of orientation by pulling. Hereditas 37:85–156
Margolis RL, Wilson L (1978) Opposite end assembly and disassembly of microtubules at steady state in vitro. Cell 13:1–8
Subtelny SS, Green PB, Society for Developmental Biology (1982) Developmental order, its origin and regulation. Liss, New York
Hill TL (1985) Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA 82:4404–4408
Joglekar AP, Hunt AJ (2002) A simple, mechanistic model for directional instability during mitotic chromosome movements. Biophys J 83:42–58
Waters JC, Mitchison TJ, Rieder CL, Salmon ED (1996) The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell 7:1547–1558
Miyamoto DT, Perlman ZE, Burbank KS, Groen AC, Mitchison TJ (2004) The kinesin Eg5 drives poleward microtubule flux in Xenopus laevis egg extract spindles. J Cell Biol 167:813–818
Rath U, Rogers GC, Tan D, Gomez-Ferreria MA, Buster DW, Sosa HJ, Sharp DJ (2009) The Drosophila kinesin-13, KLP59D, impacts Pacman- and flux-based chromosome movement. Mol Biol Cell 20:4696–4705
Zhang D, Rogers GC, Buster DW, Sharp DJ (2007) Three microtubule severing enzymes contribute to the “Pacman–flux” machinery that moves chromosomes. J Cell Biol 177:231–242
Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117:5461–5477
Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon ED, Odde DJ (2005) Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell 16:3764–3775
Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O’Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED, Bloom K, Odde DJ (2008) Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell 135:894–906
Matos I, Pereira AJ, Lince-Faria M, Cameron LA, Salmon ED, Maiato H (2009) Synchronizing chromosome segregation by flux-dependent force equalization at kinetochores. J Cell Biol 186:11–26
Gardner MK, Odde DJ (2006) Modeling of chromosome motility during mitosis. Curr Opin Cell Biol 18:639–647
Civelekoglu-Scholey G, Sharp DJ, Mogilner A, Scholey JM (2006) Model of chromosome motility in Drosophila embryos: adaptation of a general mechanism for rapid mitosis. Biophys J 90:3966–3982
Saxton WM, McIntosh JR (1987) Interzone microtubule behavior in late anaphase and telophase spindles. J Cell Biol 105:875–886
Baskin TI, Cande WZ (1990) Kinetic analysis of mitotic spindle elongation in vitro. J Cell Sci 97:79–89
Cheerambathur DK, Civelekoglu-Scholey G, Brust-Mascher I, Sommi P, Mogilner A, Scholey JM (2007) Quantitative analysis of an anaphase B switch: predicted role for a microtubule catastrophe gradient. J Cell Biol 177:995–1004
Hogan CJ, Wein H, Wordeman L, Scholey JM, Sawin KE, Cande WZ (1993) Inhibition of anaphase spindle elongation in vitro by a peptide antibody that recognizes kinesin motor domain. Proc Natl Acad Sci USA 90:6611–6615
Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM (1999) The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J Cell Biol 144:125–138
Acknowledgments
This review is based on a lecture on “Mitotic motors and the mechanism of mitosis” that was presented on 6 March 2009 to the graduate students taking the Cell Cycle course coordinated by Professors Helder Maiato and Claudio Sunkel at the Institute of Molecular and Cell biology, University of Porto, Portugal. The authors thank the hosts for their generous hospitality. Our work on mitosis is supported by NIH grant GM 55507.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Civelekoglu-Scholey, G., Scholey, J.M. Mitotic force generators and chromosome segregation. Cell. Mol. Life Sci. 67, 2231–2250 (2010). https://doi.org/10.1007/s00018-010-0326-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-010-0326-6