Abstract
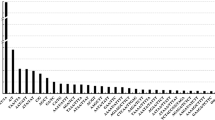
To increase the speed and reduce the cost of constructing a genetic map of Actinidia species (kiwifruit), for use in both breeding and functional genomics programmes, we sampled microsatellites from expressed sequence tags (ESTs) to evaluate their frequency of occurrence and level of polymorphism. Perfect dinucleotide repeats were the microsatellites selected, and these were found to be numerous in both the 5′ and 3′ ends of the genes represented. The microsatellites were of various lengths, the majority being repeats with the pattern (CT) n /(GA) n . One hundred and fifty microsatellites, each with more than 10 dinucleotide repeat units, were chosen as possible markers, and when these were amplified, 93.5% were found to be polymorphic and segregating in a mapping population, with 22.6% amplifying more than one locus. Four marker categories were identified. Fully informative markers made up 27% of the total, 36.2% were female informative, 25.8% were male informative and 10% partly informative. The mapping population was an intraspecific cross in the diploid species Actinidia chinensis, with parents chosen for their diversity in fruit and plant characteristics, and for their geographical separation. Linkage was tested using the software ‘Joinmap’ and a LOD value of 3. The distribution of the EST-based markers over the linkage groups obtained appeared to be random, taking into consideration the small sample size, that the number of linkage groups (31) exceeded the chromosome number of n=29, and that a number of markers were not assigned to any group. Some microsatellite markers which amplified more than one locus mapped to separate linkage groups. According to our study in A. chinensis, EST-derived microsatellites give large numbers of possible markers very quickly and at reasonable cost. The markers are highly polymorphic, segregate in the mapping population, and increase the value of the genomic map by providing some functional information.
Similar content being viewed by others
References
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457
Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68:799–808
Callen DF, Thompson AD, Shen Y, Phillips HA, Richards RI, Mulley JC, Sutherland GR (1993) Incidence and origin of “null” alleles in the (AC)n microsatellite markers. Am J Hum Genet 52:922–927
Cho YG, Ishii T, Temnykh S, Chen X, Lipovich L, McCouch SR, Parl WD, Ayres N, Cartinhour S (2000) Diversity of microsatellites derived from genomic libraries and GenBank sequences in rice (Oryza satica L.). Theor Appl Genet 100:713–722
Crowhurst RN, Gardner RC (1991) A genomic-specific repeat sequence from kiwifruit Actinidia deliciosa var. deliciosa. Theor Appl Genet 81:71–78
Eujayl I, Sorrells ME, Baum M, Wolters P, Powell W (2002) Isolation of EST-derived microsatellite markers for genotyping the A and B genomes of wheat. Theor Appl Genet 104:399–407
Huang WG, Cipriani G, Morgante M, Testolin R (1998) Microsatellite DNA in Actinidia chinensis: isolation, characterisation, and homology in related species. Theor Appl Genet 97:1269–1278
Lehmann T, Hawley WA, Collins FH (1996) An evaluation of evolutionary constraints on microsatellite loci using null alleles. Genetics 144:1155–1163
Martienssen RA, Colot V (2001) DNA methylation and epigenetic inheritance in plants and filamentous fungi. Science 293:1070–1074
McNeilage MA, Considine JA (1989) Chromosome studies in some Actinidia taxa and implications for breeding. NZ J Bot 27:71–81
Milbourne D, Russell J (1998) Comparison of molecular marker assays in inbreeding (barley) and outbreeding (potato) species. In: Molecular tools for screening biodiversity. Chapman and Hall, London
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Nadir E, Margalit H, Gallily T, Ben-Sasson SA (1996) Microsatellites spreading in the human genome: evolutionary mechanisms and structural implications Proc Natl Acad Sci 93:6470–6475
Ramsay L, Macaulay M, Cardle L, Morgante M, degli Ivanissevich S, Maestri E, Powell W, Waugh R (1999) Intimate association of microsatellite repeats with retrotransposons and other dispersed repetitive elements in barley. Plant J 17:415–425
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana, Totowa, pp 365–386
Schlotterer C, Tautz D (1992) Slippage synthesis of simple sequence DNA. Nucleic Acids Res 20:211–215
Scott KD, Eggler P, Seaton G, Rossetto M, Ablett EM, Lee LS, Henry RJ (2000) Analysis of SSRs derived from grape ESTs. Theor Appl Genet 100:723–726
Tang S, Knapp SJ (2003) Microsatellites uncover extraordinary diversity in the native American land races and wild populations of cultivated sunflower. Theor Appl Genet 106:990–1003
Testolin R, Huang WG, Lain O, Messina R, Vecchione A, Cipriani G (2001) A kiwifruit (Actinidia spp.) linkage map based on microsatellites and integrated with AFLP markers. Theor Appl Genet 103:30-36
The Huntington’s Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983
Valdes AM, Slatkin M, Freimer NB (1993) Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genome 133:737–749
Vincent JB, Patterson AD, Strong E, Petronis A, Kennedy JL (2000) The unstable trinucleotide repeat story of major psychosis. Am J Med Genet 97:77–97
Weber JL, Polymeropoulos M, May P, Kwitek A, Xiao H, McPherson JD, Wasmuth JJ (1991) Mapping of human chromosome 5 microsatellite polymorphisms. Genomics 11:695–700
Weising K, Fung RWM, Keeling DJ, Atkinson RG, Gardner R (1996) Characterisation of microsatellites from Actinidia chinensis. Mol Breed 2:117–131
Acknowledgements
This research was partially funded by the New Zealand Foundation for Research, Science and Technology (CO6X0213) and by the HortResearch Royalty Fund.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by P. Langridge
Rights and permissions
About this article
Cite this article
Fraser, L.G., Harvey, C.F., Crowhurst, R.N. et al. EST-derived microsatellites from Actinidia species and their potential for mapping. Theor Appl Genet 108, 1010–1016 (2004). https://doi.org/10.1007/s00122-003-1517-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-003-1517-4