Abstract
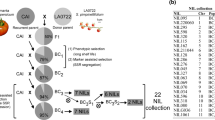
The efficiency of marker-assisted backcross for the introgression of a quantitative trait locus (QTL) from a donor line into a recipient line depends on the stability of QTL expression. QTLs for six quality traits in tomato (fruit weight, firmness, locule number, soluble solid content, sugar content and titratable acidity) were studied in order to investigate their individual effect and their stability over years, generations and genetic backgrounds. Five chromosome regions carrying fruit quality QTLs were transferred following a marker-assisted backcross scheme from a cherry tomato line into three modern lines with larger fruits. Three sets of genotypes corresponding to three generations were compared: (1) an RIL population, which contained 50% of each parental genome, (2) three BC3S1 populations which segregated simultaneously for the five regions of interest but were almost fully homozygous for the recipient genome on the eight chromosomes carrying no QTL and (3) three sets of QTL-NILs (BC3S3 lines) which differed from the recipient line only in one of the five regions. QTL detection was performed in each generation, in each genetic background and during 2 successive years for QTL-NILs. About half of the QTLs detected in QTL-NILs were detected in both years. Eight of the ten QTLs detected in RILs were recovered in the QTL-NILs with the genetic background used for the initial QTL mapping experiment, with the exception of two QTLs for fruit firmness. Several new QTLs were detected. In the two other genetic backgrounds, the number of QTLs in common with the RILs was lower, but several new QTLs were also detected in advanced generations.



Similar content being viewed by others
Abbreviations
- Bgb:
-
Vil B genetic background
- Dgb:
-
Vil D genetic background
- EUG:
-
Eugenol
- FW:
-
Fruit weight
- Lgb:
-
Levovil genetic background
- LONB:
-
Locule number
- MAS:
-
Marker-assisted selection
- MYP:
-
Orthometoxyphenol
- SSC:
-
Soluble solids content
- SUC:
-
Sugar content
- TA:
-
Titratable acidity
References
Ahmadi N, Albar L, Pressoir G, Pinel A, Fargette D, Ghesquière A (2001) Genetic basis and mapping of the resistance to Rice yellow mottle virus. III. Analysis of QTL efficiency in introgressed progenies confirmed the hypothesis of complementary epistasis between two resistance QTLs. Theor Appl Genet 103:1084–1092
Asins MJ (2002) Present and future of quantitative trait locus analysis in plant breeding. Plant Breed 121:281–291
Bouchez A, Hospital F, Causse M, Gallais A, Charcosset A (2002) Marker-assisted introgression of favorable alleles at quantitative trait loci between maize elite lines. Genetics 162:1945–1959
Causse M, Saliba-Colombani V, Lecomte L, Duffé P, Rousselle P, Buret M (2002) QTL analysis of fruit quality in fresh market tomato: a few chromosome regions control the variation of sensory and instrumental traits. J Exp Bot 53:2089–2098
Causse M, Saliba-Colombani V, Lesschaeve I, Buret M (2001) Genetic analysis of organoleptic quality in fresh market tomato. 2. Mapping QTLs for sensory attributes. Theor Appl Genet 102:273–283
Dekkers JCM, Hospital F (2002) The use of molecular genetics in the improvement of agricultural populations. Nat Rev Genet 3:22–32
Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141:1147–1162
Frary A, Nesbitt TC, Frary A, Grandillo S, Van Der Knaap E, Cong B, Liu J, Meller J, Elber R, Alpert KB, Tanksley SD (2000) fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289:85–88
Fridman E, Liu YS, Carmel-Goren L, Gur A, Shoresh M, Pleban T, Eshed Y, Zamir D (2002) Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol Genet Genomics 266:821–826
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Lander ES, Green P, Abrahamson J, Barlox A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lecomte L, Duffé P, Buret M, Hospital F, Causse M (2004a) Marker-assisted introgression of five QTLs controlling fruit quality traits into three tomato lines revealed interactions between QTLs and genetic backgrounds. Theor Appl Genet 109:658–668
Lecomte L, Saliba-Colombani V, Gautier A, Gomez-Jimenez MC, Duffé P, Buret M, Causse M (2004b) Fine mapping of QTLs of chromosome 2 affecting the fruit architecture and composition of tomato. Mol Breed 13:1–14
Li Z, Jakkula L, Hussey RS, Tamulonis JP, Boerma HR (2001) SSR mapping and confirmation of the QTL from PI96354 conditioning soybean resistance to southern root-knot nematode. Theor Appl Genet 103:1167–1173
Lincoln SE, Daly MJ, Lander ES (1992) Constructing genetic maps with MAPMAKER/EXP version 3.0. Whitehead Institute Technical Report
Lippman Z, Tanksley SD (2001) Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 158:413–422
Mohan M, Nair S, Bhagwat A, Krishna TG, Yano M, Bhatia CR, Sasaki T (1997) Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol Breed 3:87–103
Monforte AJ, Tanksley SD (2000) Fine mapping of a quantitative trait locus (QTL) from Lycopersicon hirsutum chromosome 1 affecting fruit characteristics and agronomic traits: breaking linkage among QTLs affecting different traits and dissection of heterosis for yield. Theor Appl Genet 100:471–479
Reyna N, Sneller CH (2001) Evaluation of marker-assisted introgression of yield QTL alleles into adapted soybean. Crop Sci 41:1317–1321
Robert VJM, West MAL, Inai S, Caines A, Arntzen L, Smith JK, St Clair DA (2001) Marker-assisted introgression of blackmold resistance QTL alleles from wild Lycopersicon cheesmanii to cultivated tomato (L. esculentum) and evaluation of QTL phenotypic effects. Mol Breed 8:217–233
Romagosa I, Han F, Ullrich SE, Hayes PM, Wesenberg DM (1999) Verification of yield QTL through realized molecular marker-assisted selection responses in a barley cross. Mol Breed 5:143–152
Saliba-Colombani V, Causse M, Gervais L, Philouze J (2000) Efficiency of AFLP, RAPD and RFLP markers for the construction of an intraspecific map of the tomato genome. Genome 43:29–40
Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M (2001) Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor Appl Genet 102:259–272
SAS Institute (1988) SAS users guide: statistics, SAS Institute, Cary North Carolina (USA)
Sebolt AM, Shoemaker RC, Diers BW (2000) Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci 40:1438–1444
Servin B, Dillmann C, Decoux G, Hospital F (2002) MDM: a program to compute fully informative genotype frequencies in complex breeding schemes. J Hered 93:227–228
Shen L, Courtois B, McNally KL, Robin S, Li Z (2001) Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor Appl Genet 103:75–83
Stuber CW, Sisco PH (1992) Marker-facilitated transfer of QTL alleles between elite inbred lines and responses in hybrids. In: 46th annual corn and sorghum research conference, American Seed Trade Assoc Ed, pp 104–113
Tanksley SD (1993) Mapping polygenes. Annu Rev Genet 27:205–233
Tanksley SD, Nelson JC (1996) Advanced backcross QTL analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Toojinda T, Baird E, Booth A, Broers L, Hayes P, Powell W, Thomas W, Vivar H, Young G (1998) Introgression of quantitative trait loci (QTLs) determining stripe rust resistance in barley: an example of marker-assisted line development. Theor Appl Genet 96:123–131
Van Berloo R, Aalbers H, Werkman A, Niks RE (2001) Resistance QTL confirmed through development of QTL-NILs for barley leaf rust resistance. Mol Breed 8:187–195
Yousef GG, Juvik JA (2002) Enhancement of seedling emergence in sweet corn by marker-assisted backcrossing of beneficial QTL. Crop Sci 42:96–104
Zeng Z B (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhu H, Briceño G, Dovel R, Hayes PM, Liu BH, Liu CT, Ullrich SE (1999) Molecular breeding for grain yield in barley: an evaluation of QTL effects in a spring barley cross. Theor Appl Genet 98:772–779
Acknowledgements
Many thanks to N. Baffert, A. Gautier and A. Luciani for genotypic and phenotypic works, to A.M. Cossalter for taking care of the plants and for her help during the phenotypic evaluations, to P. Duffé for plant genotyping, to R. Matthieu for chemical evaluations, to C. Dillmann for SAS programs and to Rebecca Stevens for English corrections. Laurent Lecomte was partly supported by the Conseil Régional Provence-Alpes-Côte d’Azur (France). The experiments comply with the current French laws.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by I. Paran
Rights and permissions
About this article
Cite this article
Chaïb, J., Lecomte, L., Buret, M. et al. Stability over genetic backgrounds, generations and years of quantitative trait locus (QTLs) for organoleptic quality in tomato. Theor Appl Genet 112, 934–944 (2006). https://doi.org/10.1007/s00122-005-0197-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-005-0197-7