Abstract
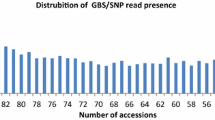
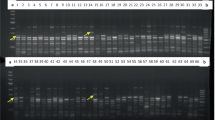
Understanding the distribution of genetic diversity among individuals, populations and gene pools is crucial for the efficient management of germplasm collections and breeding programs. Diversity analysis is routinely carried out using sequencing of selected gene(s) or molecular marker technologies. Here we report on the development of Diversity Arrays Technology (DArT) for pigeonpea (Cajanus cajan) and its wild relatives. DArT tests thousands of genomic loci for polymorphism and provides the binary scores for hundreds of markers in a single hybridization-based assay. We tested eight complexity reduction methods using various combinations of restriction enzymes and selected PstI/HaeIII genomic representation with the largest frequency of polymorphic clones (19.8%) to produce genotyping arrays. The performance of the PstI/HaeIII array was evaluated by typing 96 accessions representing nearly 20 species of Cajanus. A total of nearly 700 markers were identified with the average call rate of 96.0% and the scoring reproducibility of 99.7%. DArT markers revealed genetic relationships among the accessions consistent with the available information and systematic classification. Most of the diversity was among the wild relatives of pigeonpea or between the wild species and the cultivated C. cajan. Only 64 markers were polymorphic among the cultivated accessions. Such narrow genetic base is likely to represent a serious impediment to breeding progress in pigeonpea. Our study shows that DArT can be effectively applied in molecular systematics and biodiversity studies.


Similar content being viewed by others
References
Anderson M (2003) PCO: a FORTRAN computer program for principal coordinate analysis. University of Auckland, New Zealand
Anderson JA, Churchill G.A, Autrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Berger J, Abbo S, Turner NC (2003) Ecogeography of annual wild Cicer species: the poor state of the world collection. Crop Sci 43:1076–1090
Botstein D, Skolnick M, Davis R (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314–331
Felsenstein J (1989) PHYLIP—phylogeny inference package (Version 3.2). Cladistics 164–166
Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) version 3.6. University of Washington, Seattle
Frary A, Fulton TM, Zamir D, Tanksley SD (2004) Advanced backcross QTL analysis of a Lycopersicon esculentum × L. pennellii cross and identification of possible orthologs in the Solanaceae. Theor Appl Genet 108:485–96
Gonzalez J (1993) Random amplified polymorphic DNA analysis in Hordeum species. Genome 36:1029–1031
Huang XQ, Coster H, Ganal MW, Roder MS (2003) Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (Triticum aestivum L.). Theor Appl Genet 106:1379–89
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29(4):e25
Jain A, Bhatia S, Banga S, Lakshmikumaran M (1994) Potential use of random amplified polymorphic DNA (RAPD) technique to study the genetic diversity in Indian mustard (Brassica juncea) and its relationship to heterosis. Theor Appl Genet 88:116–122
Kumar S, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
van der Maesen LJG (1990) Pigeonpea: origin, history, evolution, and taxonomy. In: Nene YL, Hill SH, Sheila VK (eds) The pigeonpea. CABInternational, Wellingford, UK, P15–46
Mackill D (1995) Classifying Japonica rice cultivars with RAPD markers. Crop Sci 35:889–894
Miller J (1990) RFLP analysis of phylogenetic relationships and genetic variation in the genus Lycopersicon. Theor Appl Genet 80:437–448
Moretzsohn M de C, Hopkins MS, Mitchell SE, Kresovich S, Valls JF, Ferreira ME (2004) Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol 14(4):11
Ohri D, Singh S (2002) Karyotypic and genome size variation in Cajanus cajan (L.) Millsp (pigeonpea) and some wild relatives. Genet Resour Crop Evol 49:1–10
Ohri D, Jha S, Kumar S (1994) Variability in nuclear-DNA Content within Pigeonpea, Cajanus cajan (Fabaceae). Plant Syst Evol 189:211–2
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Ronald D (1999) A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep 17:53–57
Saxena K, Yang S (2000) ICRISAT Pigeonpea Jumps over Himalayas. Int Chickpea and Pigeonpea Newslett 2–3
Saxena K, Zong X, Yang S, Li Z, Zhou C (2000) Potential of Pigeonpea in China and its Genetic Improvement at ICRISAT, ICETS 2000, Beijing
Septiningsih EM, Trijatmiko KR, Moeljopawiro S, McCouch SR (2003) Identification of quantitative trait loci for grain quality in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O. rufipogon. Theor Appl Genet 107:1433–1441
Sivaramakrishnan, Reddy L (2002) Diversity in selected wild and cultivated species of pigeonpea using RFLP of mtDNA. Euphytica 125:121–128
Stein N (2001) A new DNA extraction method for high-throughput marker analysis in a large-genome species such as Triticum aestivum. Plant Breed 120:354–356
Vos P, Hoger R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Wang D, Fan J, Siao C, Berno A, Young P, Sapolsky R, Ghandour G, Perkins N, Winchester E, Spencer J et al (1998) Large-scale identification, mapping and genotyping of single-nucleotide polymorphisms in the human genome. Science 280(5366):1077–1082
Weber J, May P (1989) Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am J Hum Genet 44(3):388–396
Wenzl P, Kudrna D, Jaccoud D, Huttner E, Kleinhofs A, Kilian A (2004) Diversity arrays technology (DArT) for whole-genome profiling of barley. Proc Natl Acad Sci USA 101(26):9915–9920
Williams J, Kubelik A, Livak K, Rafalski J, Tingey S (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18(22):6531–6535
Wittenberg AH, Van der Lee T, Cayla C, Kilian A, Visser RG, Schouten HJ (2005) Validation of the high-throughput marker technology DArT using the model plant Arabidopsis thaliana. Mol Gen Genomics, 274:30–39
Xia L, Peng K, Yang S, Wenzl P, Carmen de Vicente M, Fregene M, Kilian A (2005) DArT for high-throughput genotyping of cassava (Manihot esculenta) and its wild relatives. Theor Appl Genet 110:1092–1098 DOI:101007/s100122-101005-101937-101004
Yang S, Pang W, Zong X, Li Z, Zhou C, Saxena K, Liang H (2001) A potential fodder crop for Guangxi province of China. Int Chickpea Pigeonpea Newslett 54
Acknowledgments
The authors would like to thank Dr. K. B. Saxena of ICRISAT, Patancheru, India, and Sally Dillon of ATGCRC, for providing seeds; Dr. H. D. Upadhyaya of ICRISAT, for providing pedigree information; Dr. Rongbai Li, Ms Yuanhua Chen of Guangxi Academy of Agricultural Sciences, Nanning, China and Ms Jianping Guan and Mr. Long Yuan of Chinese Academy of Agricultural Sciences, Beijing, China for helping with DNA extraction; Ms Vanessa Caig and Ms Margaret Evers, for technical help; Mr. Cyril Cayla, Mr. Grzegorz Uszynski, Dr. Yuantu Huang and Damian Jaccoud for help with data analysis. We thank all the other colleagues at DArT P/L for helpful discussions. Special thanks to Center for the Application of Molecular Biology to International Agriculture and The Finkel Family Trust for financial support for Shiying Yang.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D. A. Hoisington
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yang, S., Pang, W., Ash, G. et al. Low level of genetic diversity in cultivated Pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet 113, 585–595 (2006). https://doi.org/10.1007/s00122-006-0317-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-006-0317-z