Abstract
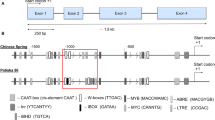
Asr (ABA, stress, ripening) genes represent a small gene family potentially involved in drought tolerance in several plant species. To analyze their interest for rice breeding for water-limited environments, this gene family was characterized further. Genomic organization of the gene family reveals six members located on four different chromosomes and with the same exon–intron structure. The maintenance of six members of the Asr gene family, which are the result of combination between tandem duplication and whole genome duplication, and their differential regulation under water stress, involves probably some sub-functionalization. The polymorphism of four members was studied in a worldwide collection of 204 accessions of Oryza sativa L. and 14 accessions of wild relatives (O. rufipogon and O. nivara). The nucleotide diversity of the Asr genes was globally low, but contrasted for the different genes, leading to different shapes of haplotype networks. Statistical tests for neutrality were used and compared to their distribution in a set of 111 reference genes spread across the genome, derived from another published study. Asr3 diversity exhibited a pattern concordant with a balancing selection at the species level and with a directional selection in the tropical japonica sub-group. This study provides a thorough description of the organization of the Asr family, and the nucleotide and haplotype diversity of four Asr in Oryzasativa species. Asr3 stood out as the best potential candidate. The polymorphism detected here represents a first step towards an association study between genetic polymorphisms of this gene family and variation in drought tolerance traits.






Similar content being viewed by others
References
Achaz G, Coissac E, Viari A, Netter P (2000) Analysis of intrachromosomal duplications in yeast Saccharomyces cerevisiae: a possible model for their origin. Mol Biol Evol 17:1268–1275
Achaz G, Netter P, Coissac E (2001) Study of intrachromosomal duplications among the eukaryote genomes. Mol Biol Evol 18:2280–2288
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148:6–24
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Bray EA (1993) Molecular responses to water deficit. Plant Physiol 103:1035–1040
Caicedo AL, Williamson SH, Hernandez RD, Boyko A, Fledel-Alon A, York TL, Polato NR, Olsen KM, Nielsen R, McCouch SR, Bustamante CD, Purugganan MD (2007) Genome-wide patterns of nucleotide polymorphism in domesticated rice. Plos Genet 3:1745–1756
Cakir B, Agasse A, Gaillard C, Saumonneau A, Delrot S, Atanassova R (2003) A grape ASR protein involved in sugar and abscisic acid signaling. Plant Cell 15:2165–2180
Caldana C, Scheible WR, Mueller-Roeber B, Ruzicic S (2007) A quantitative RT-PCR platform for high-throughput expression profiling of 2500 rice transcription factors. Plant Methods 3
Canel C, Baileyserres JN, Roose ML (1995) Pomelo fruit transcript homologous to ripening-induced genes. Plant Physiol 108:1323–1324
Carrari F, Fernie AR, Iusem ND (2004) Heard it through the grapevine? ABA and sugar cross-talk: the ASR story. Trends Plant Sci 9:57–59
Chang SJ, Puryear JD, Dias M, Funkhouser EA, Newton RJ, Cairney J (1996) Gene expression under water deficit in loblolly pine (Pinus taeda): isolation and characterization of cDNA clones. Physiol Plant 97:139–148
Chapman BA, Bowers JE, Feltus FA, Paterson AH (2006) Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc Natl Acad Sci USA 103:2730–2735
Courtois B, Lafitte R (1999) Improving rice for drought-prone upland environments. In: O’Toole J, Ito O, Hardy B (eds) Genetic improvement of rice for water-limited environments. IRRI, Los Banos, pp 35–56
de Vienne D, Leonardi A, Damerval C, Zivy M (1999) Genetics of proteome variation for QTL characterization: application to drought-stress responses in maize. J Exp Bot 50:303–309
Droc G, Ruiz M, Larmande P, Pereira A, Piffanelli P, Morel JB, Dievart A, Courtois B, Guiderdoni E, Perin C (2006) OryGenesDB: a database for rice reverse genetics. Nucleic Acids Res 34:D736–D740
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:1–19
Ehrenreich IM, Purugganan MD (2006) The molecular genetic basis of plant adaptation. Am J Bot 93:953–962
FAOSTAT (2008) Food and Agriculture Organization of the United Nations. http://faostat.fao.org/
Figueiredo LFD, Calatayud C, Dupuits C, Billot C, Rami JF, Brunel D, Perrier X, Courtois B, Deu M, Glaszmann JC (2008) Phylogeographic evidence of crop neodiversity in sorghum. Genetics 179:997–1008
Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J (1999) Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545
Frankel N, Hasson E, Iusem ND, Rossi MS (2003) Adaptive evolution of the water stress-induced gene Asr2 in Lycopersicon species dwelling in arid habitats. Mol Biol Evol 20:1955–1962
Frankel N, Carrari F, Hasson E, Iusem ND (2006) Evolutionary history of the Asr gene family. Gene 378:74–83
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Fukai S, Cooper M (1995) Development of drought-resistant cultivars using physio-morphological traits in rice. Field Crops Res 40:67–86
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Giombini MI, Frankel N, Iusem ND, Hasson E (2009) Nucleotide polymorphism in the drought responsive gene Asr2 in wild populations of tomato. Genetica 136:13–25
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 30:781–792
Holt SJ, Cress WA, Van Staden J (2002) Evidence for dynamic alteration in histone gene clusters of Caenorhabditis elegans: a topoisomerase II connection? Genet Res 79:11–22
Hudson RR, Kreitman M, Aguade M (1987) A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159
Iusem ND, Bartholomew DM, Hitz WD, Scolnik PA (1993) Tomato (Lycopersicon-Esculentum) transcript induced by water-deficit and ripening. Plant Physiol 102:1353–1354
Jeanneau M, Gerentes D, Foueillassar X, Zivy M, Vidal J, Toppan A, Perez P (2002) Improvement of drought tolerance in maize: towards the functional validation of the Zm-Asr1 gene and increase of water use efficiency by over-expressing C4-PEPC. Biochimie 84:1127–1135
Kalifa Y, Gilad A, Konrad Z, Zaccai M, Scolnik PA, Bar-Zvi D (2004) The water- and salt-stress-regulated Asr1 (abscisic acid stress ripening) gene encodes a zinc-dependent DNA-binding protein. Biochem J 381:373–378
Katju V, Lynch M (2003) The structure and early evolution of recently arisen gene duplicates in the Caenorhabditis elegans genome. Genetics 165:1793–1803
Kato S, Kosaka H, Hara S (1928) On the affinity of rice varieties as shown by the fertility of rice plants. Centr Agric Kyushu Imp Univ 2:241–276
Kim BR, Nam HY, Kim SU, Kim SI, Chang YJ (2003) Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol Lett 25:1869–1872
Konrad Z, Bar-Zvi D (2008) Synergism between the chaperone-like activity of the stress regulated ASR1 protein and the osmolyte glycine-betaine. Planta 227:1213–1219
Kovach MJ, Sweeney MT, McCouch SR (2007) New insights into the history of rice domestication. Trends Genet 23:578–587
Li CB, Zhou AL, Sang T (2006) Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara. New Phytol 170:185–193
Lin ZW, Griffith ME, Li XR, Zhu ZF, Tan LB, Fu YC, Zhang WX, Wang XK, Xie DX, Sun CQ (2007) Origin of seed shattering in rice (Oryza sativa L.). Planta 226:11–20
Lin HN, Ouyang S, Egan A, Nobuta K, Haas BJ, Zhu W, Gu X, Silva JC, Meyers BC, Buell CR (2008) Characterization of paralogous protein families in rice. BMC Plant Biol 8
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25:402–408
Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA (2006) Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc Natl Acad Sci USA 103:9578–9583
Lu BR, Zheng KL, Qian HR, Zhuang JY (2002) Genetic differentiation of wild relatives of rice as assessed by RFLP analysis. Theor Appl Genet 106:101–106
Ludlow MM, Muchow RC (1990) A critical evaluation of traits for improving crop yields in water-limited environments. Adv Agron 43:107–153
Luquet D, Clement-Vidal A, Fabre D, This D, Sonderegger N, Dingkuhn M (2008) Orchestration of transpiration, growth and carbohydrate dynamics in rice during a dry-down cycle. Funct Plant Biol 35:689–704
Meyer P (1995) Controlling and understanding transgene expression. Transgenics 13:332–337
Nei M (1987) Molecular evolutionary genetics, New York
Ohno S (1970) Evolution by gene duplication. Springer, New York
Oka HI (1988) Origin of cultivated rice. Scientific Societies Press, Japan
Olsen KM, Caicedo AL, Polato N, McClung A, McCouch S, Purugganan MD (2006) Selection under domestication: evidence for a sweep in the rice Waxy genomic region. Genetics 173:975–983
Pandey S, Behura R, Villano R, Naik D (2000) Economic cost of drought and farmers’ coping mechanisms: a study of rainfed rice in eastern India. In: Series IDP (ed), pp 1–35
Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101:9903–9908
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA get-blot analyses. Plant Physiol 133:1755–1767
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Rychlik W, Rhoads RE (1989) A computer program for choosing optimal oligonucleotides for filter hybridization, sequencing and in vitro amplification of DNA. Nucleic Acids Res 17:8543–8551
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) A proteomic approach to analyzing drought- and salt-responsiveness in rice. Field Crops Res 76:199–219
Schauser L, Wieloch W, Stougaard J (2005) Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J Mol Evol 60:229–237
Schneider A, Salamini F, Gebhardt C (1997) Expression pattern and promoter activity of the cold-regulated gene ci21A of potato. Plant Physiol 113:335–345
Second G, Ghesquière A (1995) Cartographie des introgressions réciproques entre les sous-espèces indica et japonica de riz cultivé. In: INRA (ed) Techniques et utilisation des marqueurs moléculaires, Paris, pp 83–93
Skriver K, Mundy H (1990) Gene expression in response to abscisic acid and osmotic stress. Plant Cell 2:503–512
Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400:667–671
Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, McCouch SR (2007) Global dissemination of a single mutation conferring white pericarp in rice. Plos Genet 3:1418–1424
Tajima F (1989) Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Takasaki H, Mahmood T, Matsuoka M, Matsumoto H, Komatsu S (2008) Identification and characterization of a gibberellin-regulated protein, which is ASR5, in the basal region of rice leaf sheaths. Mol Genet Genomics 279:359–370
Vaidyanathan R, Kuruvilla S, Thomas G (1999) Characterization and expression pattern of an abscisic acid and osmotic stress responsive gene from rice. Plant Sci 140:21–30
Vaughan DA, Lu BR, Tomooka N (2008) The evolving story of rice evolution. Plant Sci 174:394–408
Vitte C, Ishii T, Lamy F, Brar D, Panaud O (2004) Genomic paleontology provides evidence for two distinct origins of Asian rice (Oryza sativa L.). Mol Genet Genomics 272:504–511
Walsh B (2008) Using molecular markers for detecting domestication, improvement, and adaptation genes. Euphytica 161:1–17
Wang HJ, Hsu CM, Jauh GY, Wang CS (2005) A lily pollen ASR protein localizes to both cytoplasm and nuclei requiring a nuclear localization signal. Physiol Plant 123:314–320
Wang WK, Schaal BA, Chiou YM, Murakami N, Ge XJ, Huang CC, Chiang TY (2007) Diverse selective modes among orthologs/paralogs of the chalcone synthase (Chs) gene family of Arabidopsis thaliana and its relative A-halleri ssp. gemmifera. Mol Phylogenet Evol 44:503–520
Watterson GA (1975) Number of segregating sites in genetic models without recombination. Theor Popul Biol 7:256–276
Widawsky DA, O’Toole JC (1990) Prioritizing the rice biotechnology research agenda for eastern India, New York
Wright SI, Gaut BS (2005) Molecular population genetics and the search for adaptive evolution in plants. Mol Biol Evol 22:506–519
Xu SQ, Clark T, Zheng HK, Vang S, Li RQ, Wong GKS, Wang J, Zheng XG (2008) Gene conversion in the rice genome. BMC Genomics 9
Yamanaka S, Nakamura I, Nakai H, Sato YI (2003) Dual origin of the cultivated rice based on molecular markers of newly collected annual and perennial strains of wild rice species, Oryza nivara and O. rufipogon. Genet Resour Crop Evol 50:529–538
Yang CY, Chen YC, Jauh GY, Wang CS (2005) A lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in Arabidopsis. Plant Physiol 139:836–846
Yu J, Wang J, Lin W, Li SG, Li H, Zhou J et al (2005) The genomes of Oryza sativa: a history of duplications. Plos Biol 3:266–281
Zhu Q, Ge S (2005) Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol 167:632–632
Zhu QH, Zheng XM, Luo JC, Gaut BS, Ge S (2007) Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: severe bottleneck during domestication of rice. Mol Biol Evol 24:875–888
Acknowledgments
This work was funded by the Generation Challenge Program project number 2006-02. Thanks to Carine Charron and Nabila Yahiaoui for their critical reviews.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wissuwa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Philippe, R., Courtois, B., McNally, K.L. et al. Structure, allelic diversity and selection of Asr genes, candidate for drought tolerance, in Oryza sativa L. and wild relatives. Theor Appl Genet 121, 769–787 (2010). https://doi.org/10.1007/s00122-010-1348-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1348-z