Abstract
Aims/hypothesis
Complex changes in gene expression are associated with insulin resistance and non-alcoholic fatty liver disease (NAFLD) promoted by feeding a high-fat diet (HFD). We used functional genomic technologies to document molecular mechanisms associated with diet-induced NAFLD.
Materials and Methods
Male 129S6 mice were fed a diet containing 40% fat (high-fat diet, HFD) for 15 weeks. Glucose tolerance, in vivo insulin secretion, plasma lipid profile and adiposity were determined. Plasma metabonomics and liver transcriptomics were used to identify changes in gene expression associated with HFD-induced NAFLD.
Results
In HFD-fed mice, NAFLD and impaired glucose and lipid homeostasis were associated with increased hepatic transcription of genes involved in fatty acid uptake, intracellular transport, modification and elongation, whilst genes involved in beta-oxidation and lipoprotein secretion were, paradoxically, also upregulated. NAFLD developed despite strong and sustained downregulation of transcription of the gene encoding stearoyl-coenzyme A desaturase 1 (Scd1) and uncoordinated regulation of transcription of Scd1 and the gene encoding sterol regulatory element binding factor 1c (Srebf1c) transcription. Inflammatory mechanisms appeared to be stimulated by HFD.
Conclusions/interpretation
Our results provide an accurate representation of subtle changes in metabolic and gene expression regulation underlying disease-promoting and compensatory mechanisms, collectively contributing to diet-induced insulin resistance and NAFLD. They suggest that proposed models of NAFLD pathogenesis can be enriched with novel diet-reactive genes and disease mechanisms.
Similar content being viewed by others
Introduction
The increasing incidence of type 2 diabetes mellitus and obesity is paralleled by a progression in a wide spectrum of clinical conditions often associated with insulin resistance. Non-alcoholic fatty liver disease (NAFLD) is the most common liver disease associated with insulin resistance [1, 2]. Genetic and environmental factors, including dietary excess and inadequate exercise, contribute to NAFLD either directly or indirectly through inter-organ hormonal or metabolic signalling events [3, 4].
A composite description of molecular mechanisms associated with liver triacylglycerol accumulation, e.g. increased fatty acid uptake and synthesis and reduced fatty acid beta-oxidation and export, is emerging from clinical and experimental data [5]. Progress in gene expression profiling technologies and genome functional annotations can help improve knowledge of NAFLD-associated markers. Functional genomic tools, including transcriptomics and metabonomics, a technology designed to quantify metabolites in biological samples [6], generate high-density data sets underlying disease-related mechanisms at either end of the regulation of gene expression. Alterations in transcription regulation of hepatic carbohydrate and lipogenic pathways have a central role in mechanisms contributing to insulin resistance and NAFLD [7, 8].
Owing to the complex aetiology and pathogenesis of diabetes and obesity and ethical issues in relation to genomic investigations in patients and controls, rodent models are important tools for studying human disease conditions in controlled genetic and environmental situations [9]. Feeding rodents a high-fat diet (HFD) provides an efficient system for consistently promoting obesity, insulin resistance and NAFLD in the vast majority of strains [10, 11], even after short periods of fat feeding [12]. We and others have shown that 129S6 mice are susceptible to HFD-induced diabetes, obesity and fatty liver and that the molecular mechanisms involved are multifaceted [13, 14].
Here we report the detailed characterisation of physiological, metabolic and liver transcriptional responses to prolonged HFD feeding in 129S6 mice. Upregulated expression of genes involved in fatty acid uptake, intracellular transport, modification and elongation in response to HFD contributes to NAFLD despite enhanced expression of genes involved in beta-oxidation and lipoprotein secretion. These results indicate that mice susceptible to experimentally induced obesity and NAFLD adapt to HFD feeding with a complex pattern of transcriptional changes, which probably involve disease-causative mechanisms and secondary hepatic adaptations to obesity, glucose intolerance, hyperinsulinaemia and hyperlipidaemia.
Materials and Methods
Animals
Male mice from the 129S6/SvEv and BALB/c inbred strains were used. Mice were weaned at 3 weeks of age and housed in groups of ten. Mice were maintained on a 12-h light and dark cycle and fed ad libitum a standard carbohydrate (CD) chow (ERB, Witham, UK) from weaning. At 5 weeks of age, one group of mice was transferred to an HFD (Special Diets Services, Witham, UK), containing 32% lard oil and 8% corn oil. A separate age-matched group remained on the CD. All procedures were carried out in accordance with national license conditions.
Intraperitoneal glucose tolerance test
At 5 months 129S6 mice were fasted overnight and subjected to a simplified intraperitoneal glucose tolerance test (IPGTT) as previously described [15]. Mice were anaesthetised by i.p. injection of sodium pentobarbital (Sagatal; Rhône Mérieux, Harlow, UK) and injected i.p. with a single dose of 2 g glucose/kg body weight. Blood samples were collected before glucose injection and 15, 30 and 75 min afterwards. Blood glucose concentrations were determined immediately using a glucose meter (Accucheck; Roche Diagnostics, Welwyn Garden City, UK). Blood samples were centrifuged and plasma was collected for insulin assays. Cumulative glycaemia (CumG) and insulinaemia (CumIRI; IRI is immunoreactive insulin) were calculated as the increment of the values of plasma glucose and insulin, respectively, during the IPGTT. Glucose-induced insulin secretion was calculated as the insulinogenic index (ΔIRI/ΔG), which is the ratio of incremental plasma insulin values above baseline, integrated over 75 min after glucose injection (ΔIRI), to the corresponding incremental plasma glucose values (ΔG) integrated over the same period.
Tissue sampling
Mice were allowed to recover for 4 days after the IPGTT. They were separately housed in metabolic cages for one night to determine food consumption. Body weight (BW) and length (BL) (distance from the tip of the nose to the base of the tail) were determined and BMI was calculated as BW/BL2. Following an overnight fast, mice were killed by cerebral dislocation and cardiac puncture. Blood was centrifuged and plasma was removed for lipid assay and metabonomic profiling. Heart, brown fat, and epididymal and retroperitoneal white fat pads were dissected and weighed. Adiposity index was calculated as the ratio between epididymal or retroperitoneal fat weight and body weight. Liver samples were snap-frozen in liquid nitrogen and stored at −80°C for RNA preparation. A similar procedure was applied to collect blood and organs from 129S6 mice maintained on an HFD for 3, 7 and 23 weeks, and from age-matched CD-fed controls.
Analytical assays
Plasma IRI was determined using an ELISA kit (Mercodia, Uppsala, Sweden). Plasma lipids were measured using colorimetric detection assays (ABX Diagnostics, Shefford, UK) on a COBAS MIRA clinical chemistry analyser (ABX Diagnostics).
1H-NMR spectroscopy of mouse plasma
1H-NMR spectra of individual mice were acquired at 600.22 MHz on a Bruker Avance-600 spectrometer (Bruker, Coventry, UK) using a 100 μl plasma aliquot. Standard monodimensional spectra were acquired using a selective presaturation pulse sequence for water signal suppression as described elsewhere [16]. For each acquisition, 128 transient-free induction decays (FID) were collected into 32,768 data points using a spectral width of 20 ppm. The FIDs were convoluted with an exponential weighting function corresponding to a line broadening of 0.3 Hz before Fourier transform. The acquired NMR spectra were corrected for phase and baseline distortions using in-house software and referenced to glucose α-anomeric signal (δ5.23) [16]. The 1H-NMR spectra were imported as full-spectrum data [17]. To remove the effects of variation in the suppression of the water resonance, the region (δ4.5–5.0) was discarded. The data were normalised using noise variance normalisation. The final data set was then mean-centred prior to analysis [17].
RNA preparation
Total RNA was extracted from frozen liver using Trizol reagent (Invitrogen Life Technologies, Paisley, UK) and cleaned with RNeasy columns (Qiagen, Crawley, UK). RNA concentrations and integrity were assessed using an Agilent 2100 Bioanalyser (Agilent Technologies, Waldbronn, Germany).
Array hybridisation and scanning
Gene transcription profiling was carried out using Affymetrix expression arrays U430A and B (Affymetrix UK, High Wycombe, UK), which contain 22,690 (U430A) and 22,576 (U430B) probe sets, allowing the interrogation of the transcription level of 13 250 (U430A) and 7577 (U430B) independent gene and expressed sequence tag (EST) sequences. Hybridisation was carried out following cDNA synthesis and biotin-labelled cRNA synthesis was performed using 10 μg of total RNA from each liver sample. cRNA concentrations and integrity were assessed using an Agilent 2100 Bioanalyser. Biotinylated cRNAs were fragmented and individually hybridised in an Affymetrix oven to the arrays. Washing and staining procedures were performed in an Affymetrix Fluidics Station 450. Arrays were scanned at 560 nm using an Agilent array scanner (Affymetrix UK). All microarray experiments were compliant with MIAME (Minimum Information about a Microarray Experiment) and both protocols and data are publicly available through the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress/, last accessed in June 2007) under the accession E-MEXP-893.
Statistical analysis of microarray data
Microarray data were obtained from liver samples of 12 mice (six from each group). Analysis of the Affymetrix. CEL file data was performed using the Bioconductor packages in the R language and environment. The U430A and B chips were normalised separately. The analysis of significant differences between test groups for each probe set was also performed separately for each chip data set. Gene chip data were normalised by RMA quantile normalisation [18]. We used the RMA expression index, ignoring mismatch (MM) values for background correction, even though the method may underestimate transcription ratio values. A linear model for microarray data (LIMMA) was used to calculate significant differences between groups. To correct for multiple testing, we used the false discovery rate [19], which provides a measure of the number false positives for a given t statistic level.
Pathways and biological network analyses
To assess the significance of difference in expression of gene categories between fat-fed and control mice, array-wide false discovery probability was derived from analysis of 1,000 false data sets, each created by permuting the class labels (GO or KEGG term) among the probe sets. Data are presented as significantly differentially expressed GO and KEGG gene clusters (chip-wide p < 0.05) or not significant. Ingenuity systems pathway knowledge base analysis (IPA) (Ingenuity Systems, Mountain View, CA, USA) was used to identify known biological networks and pathways associated with gene expression differences in response to HFD.
Quantitative real-time PCR
Assays were performed on a Rotor-Gene 3000 system (Corbett Research, Milton, UK) using the QuantiTect SYBR Green PCR kit (Qiagen). Analysis was performed using the standard curve method (Rotor-Gene Software 5.0.47; Corbett Research, Milton, UK). This software allowed further verification that a single PCR product signal was quantified. Gene expression was normalised against the expression of beta actin. Experiments were performed in duplicate with samples prepared from six animals per group. Statistical significance was determined by the two-tailed independent sample t test.
Statistical analysis of 1H NMR data
Orthogonal partial least squares discriminant analysis (O-PLS-DA) allows enhanced focus on diet-related variation whilst minimising other biological/analytical variation. The two experimental groups were modelled using the O-PLS algorithm, which derives from the PLS regression method, as described previously [14, 17]. The method was also applied to determine global metabolic changes derived from plasma 1H NMR data in response to short-term and prolonged fat feeding in 129S6 mice. Further details of standard O-PLS implementation in metabonomics have been given previously [17, 20].
Results
Metabolic and hormonal adaptation to prolonged fat feeding
After 15 weeks of fat feeding, 129S6 mice exhibited significantly increased body weight, BMI, adiposity indices and plasma concentration of triacylglycerol and total, HDL- and LDL-cholesterol when compared with CD-fed controls (Fig. 1). Food and digestible energy consumption was lower in HFD-fed mice than in CD-fed controls. Fat feeding induced a threefold increase in fasting insulinaemia without significant changes in fasting glycaemia (Fig. 2) and a significantly increased ratio between fasting insulin and fasting glucose (35.5 ± 8.7 in HFD-fed mice and 9.6 ± 2.4 in CD-fed mice; p = 0.03), suggesting a reduction in the biological effects of insulin in the HFD-fed group. Fat-fed mice developed glucose intolerance, as reflected by both hyperglycaemia 75 min after glucose injection and significantly higher values of cumulative glycaemia during the IPGTT than controls (Fig. 2). Fat feeding also induced enhanced insulin secretion, demonstrated by sustained hyperinsulinaemia during the IPGTT and increased indices of insulin secretion (Fig. 2). Enhanced insulin secretion without concomitant reduction in plasma glucose indicates decreased sensitivity to the biological action of insulin.
Effects of prolonged (15 weeks) HFD feeding (closed bars) on body weight (a, b), tissue weight (c–f), plasma lipid profile (g–j) and 12-h energy intake (k, l) in 129S6 mice (n = 6). Age-matched controls were fed CD (open bars; n = 6). These mice were used for gene transcription profiling. Data are presented as means ± SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs controls. BAT, brown adipose tissue; b.w., body weight; EFP, epididymal fat pad; RFP, retroperitoneal fat pad
Glucose tolerance (a–c) and glucose-stimulated insulin secretion in vivo (d–h) in 129S6 mice fed a HFD for 15 weeks (closed circles/bars; n = 6) and age-matched CD-fed controls (open circles/bars; n = 6). These mice were used for gene transcription profiling. Data are presented as means ± SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs controls
We verified that this group of fat-fed mice developed histological evidence of fatty liver (data not shown), as reported in 129S6 mice under identical experimental conditions [14].
NMR analysis of plasma metabolites
Plasma 1H NMR spectra were used to define biomarkers for diet-induced obesity, glucose intolerance and NAFLD. The metabolic adaptation to HFD was characterised by increases in plasma lactate, pyruvate, glucose, fucose, phosphatidylcholine, trimethylamine N-oxide and other methylamines (Table 1). This indicates that fat-fed mice develop hyperglycaemia, hyperlipidaemia, with all lipid resonances increased, and lactic acidosis.
Longitudinal effects of HFD on metabolic and physiological variables
To investigate the progression of some these metabolic features with prolonged fat feeding, a time-course phenotype study was performed in 129S6 mice. Metabolic changes suggested by multivariate O-PLS-DA modelling of metabonomic NMR data developed shortly after the initiation of fat feeding and remained significant throughout the remainder of the study (Fig. 3a). Most of the biomarkers in Table 1 account for the permanent separation of the two groups (data not shown). A similar pattern of phenotype evolution was observed for increased body weight and adiposity (Fig. 3b,c) and hyperlipidaemia (Fig. 3d–f), indicating the rapid and permanent effects of HFD on metabolic perturbations in 129S6 mice.
Longitudinal effects of HFD feeding (closed triangles/grey bars) on global metabonomic profile derived from plasma 1H NMR O-PLS-DA diet predictive score (a), body weight (b), adiposity (c), and plasma lipid concentration (d–f) in 129S6 mice. Data are presented as means ± SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs age-matched controls (open triangles/white bars). EFP, epididymal fat pad. The number of mice in each group is shown at the bottom of each bar
Effects of HFD on genome-wide liver transcription profiling
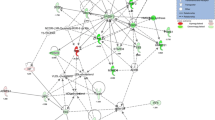
Microarray data analysis defined gene expression mechanisms underlying diet-induced impaired glucose homeostasis, obesity and NAFLD. Fat feeding induced transcriptional changes in 459 probe sets corresponding to 356 different genes and 43 apparently different EST sequences or predicted genes. These genes were considered for data biological interpretation regardless of gene expression indices (Supplementary Table 1). Interactome mapping of differentially expressed genes using IPA revealed that the high-level functions and canonical pathways most significantly affected by HFD in liver were the metabolism of tryptophan and fatty acid (Fig. 4). The urea cycle and the metabolism of amino groups pathways were predominantly affected (decreased expression).
Effects of prolonged HFD feeding on transcriptional changes of canonical pathways in 129S6 mice. Pathways significantly (p < 0.05) affected by fat feeding were identified using IPA and are ranked according to their statistical significance. Percentages of differentially expressed genes in each pathway are shown. The phenotypes of the six fat-fed and control mice used are shown in Figs. 1 and 2
At the individual gene level, the transcription of genes involved in lipoprotein uptake (Scarb1 and Cd36) was upregulated in response to HFD feeding (though Cd36 RNA levels were low; Table 2). Genes concerned with lipoprotein secretion and lipid and cholesterol efflux (Abca1, Abcb11, Apoa1, Cyp7b1, Saa4), fatty acid beta-oxidation (Acat1), intracellular fatty acid transport (Fabp5) and fatty acid and sterol synthesis, modification and elongation (Srebf1c [also known as Srebf1], Acsl5, Elovl3, Fads2, Hsd17b12, Sc5d) were upregulated by HFD. In contrast, transcript abundance of Scd1, the gene encoding the enzyme stearoyl-coenzyme A desaturase 1, which catalyses a rate-limiting step in the synthesis of unsaturated fatty acids, was very significantly reduced in response to HFD. The expression of a series of n-3 polyunsaturated fatty acid (PUFA) epoxygenases (Cyp2c29, Cyp2c37, Cyp2c38, Cyp2c39) was markedly suppressed by HFD.
In fat-fed mice, significant changes in the abundance of transcripts for Aldo2 (−20%), Gpd2 (up to +90%) and Gyk (up to +40%) may drive metabolism to increased production of glycerol-3-phosphate, the key substrate for triacylglycerol synthesis, whilst parallel downregulation of Mgll expression may limit the degradation of intracellular triacylglycerol stores. The transcription of enzymes of the urea cycle (Arg1, Asl, Ass1) was reduced by HFD. No major changes in transcripts for enzymes involved in carbohydrate metabolism were noted. These data suggest that increased lipoprotein uptake, intracellular fatty acid transport and elongation in response to HFD may account for the NAFLD phenotype, despite apparent reduced de novo lipid biosynthesis and enhanced lipid and cholesterol efflux.
Adaptation to HFD feeding also involved changes in expression of genes associated with inflammation (increased C4bp, C6, Ikbkg, Lifr, Tgfbi; decreased C5ar1, Serpinb5, Mapk11), DNA damage (increased Trp53inp2), cell death (increased Casp8, decreased Mapk11), increased production of cellular reactive oxygen species (ROS) (Nox4) and decreased chaperones and free radical scavengers (Mt1) (Table 2).
In addition, expression of several genes encoding glutathione S-transferases (Gsta1, Gsta2, Gstm1, Gsto1, Gstt2, Gstt3), an aldo-keto reductase (Akr1c19) and an aldehyde dehydrogenase (Aldh1b1) was downregulated by HFD, suggesting reduced glutathione conjugation and resulting in reduced ROS deactivation processes and increased proinflammatory activities. These patterns of gene expression suggest further deterioration of liver histopathological features in fat-fed mice.
Gene transcription regulation assessed by quantitative real-time PCR
Quantitative real-time PCR was used to detect possible false positive and false negative results from microarray analysis for key genes biologically relevant to insulin resistance and NAFLD (Figs. 5 and 6), selected on the basis of differential expression between fat fed mice and controls (Acat1, Adfp, Ass1, Cyp2c29, Fabp5, Scd1, Srebf1c) in the Affymetrix data set, lack of statistically significant response to HFD (Acox1, Acsl1, Fabp1, Onecut1 [also known as Hnf6], Ppara) despite 20–40% fold changes in transcription, or inconclusive results from different probe sets (Gyk). Assays replicated the significance and direction of transcriptional changes found by microarray screening for Ass1, Cyp2c29, Fabp5, Scd1 and Srebf1c, but failed to confirm gene expression changes for Acat1, Adfp and Gyk (probe set 1422704_at). We confirmed the absence of effect of HFD on the expression of Acsl1, Fabp1 and Gyk (probe set 1422703_at). In contrast, the transcription of Acox1, Onecut1 and Ppara was significantly downregulated by HFD when tested by quantitative real-time PCR. Differences in target sequences used in the two systems may explain these discordant results.
Effects of prolonged (15 weeks) HFD feeding on gene transcription regulation analysed by quantitative real-time PCR in 129S6 (a) and BALB/c (b) mice. For each gene Affymetrix-based transcription ratios between HFD-fed (closed bars) and control (open bars) 129S6 mice are reported above quantitative real-time PCR data. Genes showing significant differential expression on the Affymetrix data set are underlined. Details of the oligonucleotide sequences and the gene regions targeted by quantitative real-time PCR analysis and Affymetrix probe sets are given in ESM Table 2. Experiments were performed in duplicate with samples prepared from six mice. The phenotypes of the six HFD- and six CD-fed 129S6 mice are shown in Figs. 1 and 2. Data are expressed as means ± SE. *p < 0.05 vs controls
Longitudinal effects of HFD feeding on the abundance of Scd1 (a), Srebf1c (b) and Ppara (c) liver transcripts in 129S6 mice. Quantitative RT-PCR was used to determine liver transcript abundance in mice fed a HFD (closed bars) for 3, 7, 15 or 23 weeks and age-matched controls fed a CD (open bars). Oligonucleotide sequences are given in ESM Table 2. Experiments were carried out in duplicate using samples of six mice selected among the animals tested for the phenotypes shown in Fig. 3. Data are expressed as means ± SE. *p < 0.05, **p < 0.01, ***p < 0.001 vs controls
To test an effect of genetic background differences on gene transcription regulation, quantitative real-time PCR was repeated in fat-fed and control BALB/c mice, which are resistant to diet-induced obesity, diabetes and fatty liver [14]. The two strains showed markedly contrasting gene transcription patterns induced by HFD, suggesting the involvement of strain-specific mechanisms of gene expression regulation underlying susceptibility or resistance to diet-induced obesity and NAFLD.
Longitudinal effects of HFD on Scd1, Srebf1c and Ppara gene transcription
We then investigated shorter- (3 and 7 weeks) and longer-term (23 weeks) effects of HFD in 129S6 mice on key genes regulating the transcription of fatty acid metabolism (Srebf1c, Ppara) and tested the paradoxical downregulation of Scd1 transcription in a model of obesity. These experiments were carried out with fat-fed mice that showed glucose intolerance (data not shown), extensive metabolic changes and increased adiposity (Fig. 3) when compared with CD fed controls and marked NAFLD after prolonged HFD feeding (23 weeks; data not shown). Gene expression data demonstrate the permanent effects of HFD on Scd1 downregulation and Srebf1c upregulation, even though the change in Srebf1c transcription at the last time point was not significant (Fig. 6). The abundance of Ppara transcripts was also reduced by HFD, but the effect was significant only in mice fat-fed for 15 weeks.
Discussion
We have used global functional genomic approaches to document the impact of dietary changes on the regulation of gene expression in an inbred mouse strain susceptible to HFD-induced obesity, diabetes and NAFLD [13, 14]. Metabonomic and transcriptomic data provide an original representation of subtle adaptations of gene expression regulatory systems to sustained and standardised dietary changes and underline the involvement of a wide range of known and potentially novel molecular mechanisms.
Pathophysiological responses to prolonged HFD in 129S6 mice combine glucose intolerance, hyperinsulinaemia, enhanced insulin secretion, hyperlipidaemia, obesity and NAFLD. Most of these features, which develop soon after the start of an HFD and remain permanently altered, are consistently found in similar fat-feeding studies in this [13, 14] and other mouse strains [11, 13, 21]. They are the result of complex metabolic and hormonal alterations that plasma metabonomics and liver transcriptomics can characterise at the molecular level. Metabolic and gene transcription changes underlie primary causes of liver fat accumulation and impaired glucose regulation induced by HFD, secondary physiological adaptations to perturbed metabolism, hyperinsulinaemia and liver morphological alterations, and diet-specific transitions between fed and fasting states.
Metabonomic data reported here provide detailed analysis of quantitative changes in individual plasma metabolites that are contributed to by multiple organs, each having specific effects on the regulation of lipid, protein and carbohydrate fuel physiology, and showing specific responses to hormonal and neural stimuli. For example, elevated plasma lactate concentration in fat-fed mice may reflect the effect of hyperglycaemia on enhanced muscle lactate production. High concentrations of plasma lactate, pyruvate and alanine in fasted mice of the fat-fed group indicate increased availability of gluconeogenic substrates. Some diet-induced changes in metabolite concentrations can be linked to alterations in liver gene expression regulation. In fat-fed mice, elevated plasma alanine, which is used for the transport of nitrogen for excretion as urea in the liver, may be caused by the downregulation of genes encoding enzymes of the urea cycle. Increased plasma n-3 fatty acids and PUFA classes in HFD-fed mice may reflect the transcription downregulation of liver epoxygenases, resulting in PUFA-sparing function. The definition of coordinated changes in plasma metabolites and gene expression could help test causal relationships between insulin resistance and NAFLD.
Hepatic insulin- and glucose-responsive metabolic systems play a central role in the regulation of carbohydrate and lipid metabolism [22]. Interpretation of gene expression profiling data relies on mechanistic models integrating molecular markers of disease onset and progression, which derive from clinical and experimental observations. A consensus model was proposed for NAFLD [5] in which hepatic lipid accumulation is promoted by increased fatty acid uptake and biosynthesis, and decreased degradation and secretion. The gene expression data reported here suggest that the tendency of 129S6 mice to develop NAFLD stems from the activation of genes for lipoprotein assembly and is compensated by an increase in the expression of genes involved in beta-oxidation. Apparent insulin resistance in 129S6 fat-fed mice would indicate accrued NEFA fluxes from peripheral tissues to the liver. This is supported by the activation of genes for cytoplasmic fatty acid binding and transport proteins and transporters for the movement of fatty acids into liver cells and organelles. In this model, however, de novo fatty acid synthesis is depressed, but there is activation of mechanisms for fatty acid modification and elongation of pre-existing fatty acids.
Our gene transcription data confirm a number of reports on the regulation of hepatic gene expression [7, 8], derived from different microarray platforms generally allowing the interrogation of largely overlapping sets of transcripts [10, 23]. In particular, stimulated Srebf1c transcription in response to short-term and prolonged fat feeding in 129S6 mice has been reported in models of spontaneous and HFD-induced insulin resistance and NAFLD [13, 24, 25]. SREBF1c is a transcription factor involved in carbohydrate and lipid metabolism [8]. Its activation, which simulates lipogenic enzymes in the liver, is mediated by increased glucose and insulin concentrations [22, 26]. In fat-fed 129S6 mice, hyperglycaemia and hyperinsulinaemia could directly result in the upregulation of Srebf1c transcription, which can in turn enhance the transcription of Abca1, encoding an Apo-A1 binding protein controlling cholesterol efflux [27].
Insulin-mediated stimulation of Srebf1c expression also contributes to the upregulation of SCD1, the rate-limiting enzyme catalysing fatty acid synthesis [28]. There is strong evidence that downregulation of SCD1 prevents or reduces the development of NAFLD, obesity and insulin resistance [29–32]. Elevated hepatic transcription of Scd1 has been reported in fat-fed mice of the 129S6 [13] and C57BL/6J [13, 23] strains. This is in sharp contrast to our results, which showed strong and permanent HFD-induced downregulation of Scd1 transcription, of similar magnitude to that previously reported in fat-fed rats [33]. Differences in experimental protocols (e.g. HFD composition, duration of fat feeding) and genetic variation between strains can explain these discrepancies. We sequenced in the C57BL6J and 129S6 strains about 2 kb of the promoter and all exons and intron-flanking regions of the gene Scd1 (GenBank NM_009127) and found no variants that can explain altered Scd1 transcription regulation in 129S6 mice (data not shown). Although this effect may underlie NAFLD compensatory mechanisms, HFD-stimulated transcription of both Srebf1c and Abca1, a protein normally destabilised and downregulated by SCD1 [27], suggests that SCD1 inhibition may involve a factor uncoupling the coordinated regulation of Srebf1c and Scd1 transcription. This may be caused by coding variants in genes which are not differentially transcribed between fat-fed and control mice, including MLX interacting protein-like (Mlxipl, also known as ChREBP) and the nuclear receptor 1H3 (Nr1h3, also known as LXR-a).
The upregulation of Srebf1c transcription in 129S6 fat-fed mice is also relevant to the role of gut microbiota in NAFLD and insulin resistance [14]. Srebf1c is overexpressed in experiments on the microbial colonisation of the gastrointestinal tract in germ-free mice [34]. Gut microbiota could promote storage of triacylglycerol in hepatocytes and adipocytes by indirect modulation of cellular fatty acid uptake [14, 34]. There is growing evidence from studies in animals [35] and humans [36] that variation in gut microbial composition is an important factor in obesity and insulin resistance. The increased level of microbial-related metabolites (dimethylamine, trimethylamine, trimethylamine N-oxide) in the plasma of 129S6 fat-fed mice indicates important changes in gut microbiota that could contribute to NAFLD, obesity and insulin resistance. This model provides a useful system to test causal links between changes in gut microbiota and these pathologies.
We show here detailed changes in gene transcription and metabolic regulation that favour NAFLD and possibly subsequent phenotype progression involving inflammatory processes in 129S6 mice, despite activation of compensatory homeostatic mechanisms. Our results help establish consensus and strain-specific patterns of gene expression associated with NAFLD, obesity and diabetes, and identify gene targets for validation studies at the proteome level [37]. With the emergence of transcriptomic data sets derived from NAFLD-susceptible strains [10, 13, 23], our results contribute to enrichment of the model proposed for disease pathogenesis [5] with novel insights into mechanisms regulating gene expression and novel diet-reactive molecular components, including predicted or poorly annotated genes. Further investigations in a genetic cross between 129S6 and a strain resistant to diet-induced NAFLD, obesity and diabetes are required to investigate causal relationships between altered transcriptomic and metabonomic patterns and disease traits [38].
Abbreviations
- CD:
-
carbohydrate diet
- EST:
-
expressed sequence tag
- HFD:
-
high-fat diet
- IPA:
-
ingenuity systems pathway knowledge base analysis
- IPGTT:
-
intraperitoneal glucose tolerance test
- IRI:
-
immunoreactive insulin
- NAFLD:
-
non-alcoholic fatty liver disease
- PUFA:
-
polyunsaturated fatty acid
- SCD1:
-
stearoyl-coenzyme A desaturase 1
- SREBF1C:
-
sterol regulatory element binding factor 1c
References
Roden M (2006) Mechanisms of disease: hepatic steatosis in type 2 diabetes—pathogenesis and clinical relevance. Nat Clin Pract Endocrinol Metab 2:335–348
Marchesini G, Brizi M, Bianchi G et al (2001) Nonalcoholic fatty liver disease: a feature of the metabolic syndrome. Diabetes 50:1844–1850
Chitturi S, Farrell GC (2001) Etiopathogenesis of nonalcoholic steatohepatitis. Semin Liver Dis 21:27–41
Festi D, Colecchia A, Sacco T, Bondi M, Roda E, Marchesini G (2004) Hepatic steatosis in obese patients: clinical aspects and prognostic significance. Obes Rev 5:27–42
Browning JD, Horton JD (2004) Molecular mediators of hepatic steatosis and liver injury. J Clin Invest 114:147–152
Nicholson JK, Wilson ID (2003) Opinion: understanding ‘global’ systems biology: metabonomics and the continuum of metabolism. Nat Rev Drug Discov 2:668–676
Desvergne B, Michalik L, Wahli W (2006) Transcriptional regulation of metabolism. Physiol Rev 86:465–514
Foufelle F, Ferre P (2002) New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. Biochem J 366:377–391
den Boer M, Voshol PJ, Kuipers F, Havekes LM, Romijn JA (2004) Hepatic steatosis: a mediator of the metabolic syndrome. Lessons from animal models. Arterioscler Thromb Vasc Biol 24:644–649
Almind K, Kahn CR (2004) Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes 53:3274–3285
Rossmeisl M, Rim JS, Koza RA, Kozak LP (2003) Variation in type 2 diabetes-related traits in mouse strains susceptible to diet-induced obesity. Diabetes 52:1958–1966
Samuel VT, Liu ZX, Qu X et al (2004) Mechanism of hepatic insulin resistance in non-alcoholic fatty liver disease. J Biol Chem 279:32345–32353
Biddinger SB, Almind K, Miyazaki M, Kokkotou E, Ntambi JM, Kahn CR (2005) Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes 54:1314–1323
Dumas ME, Barton RH, Toye A et al (2006) Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci USA 103:12511–12516
Solberg LC, Valdar W, Gauguier D et al (2006) A protocol for high-throughput phenotyping, suitable for quantitative trait analysis in mice. Mamm Genome 17:129–146
Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC (1995) 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem 67:793–811
Cloarec O, Dumas ME, Trygg J et al (2005) Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal Chem 77:517–526
Irizarry RA, Hobbs B, Collin F et al (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Trygg J, Wold S (2003) O2-PLS, a two-block (X–Y) latent variable regression (LVR) method with an integral OSC filter. J Chemometrics 17:53–64
West DB, Boozer CN, Moody DL, Atkinson RL (1992) Dietary obesity in nine inbred mouse strains. Am J Physiol 262:R1025–R1032
Weickert MO, Pfeiffer AF (2006) Signalling mechanisms linking hepatic glucose and lipid metabolism. Diabetologia 49:1732–1741
de Fourmestraux V, Neubauer H, Poussin C et al (2004) Transcript profiling suggests that differential metabolic adaptation of mice to a high fat diet is associated with changes in liver to muscle lipid fluxes. J Biol Chem 279:50743–50753
Shimomura I, Bashmakov Y, Horton JD (1999) Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J Biol Chem 274:30028–30032
Shimomura I, Matsuda M, Hammer RE, Bashmakov Y, Brown MS, Goldstein JL (2000) Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol Cell 6:77–86
Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL (1999) Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA 96:13656–13661
Wang Y, Kurdi-Haidar B, Oram JF (2004) LXR-mediated activation of macrophage stearoyl-CoA desaturase generates unsaturated fatty acids that destabilize ABCA1. J Lipid Res 45:972–980
Dobrzyn A, Ntambi JM (2004) The role of stearoyl-CoA desaturase in body weight regulation. Trends Cardiovasc Med 14:77–81
Cohen P, Miyazaki M, Socci ND et al (2002) Role for stearoyl-CoA desaturase-1 in leptin-mediated weight loss. Science 297:240–243
Yahagi N, Shimano H, Hasty AH et al (2002) Absence of sterol regulatory element-binding protein-1 (SREBP-1) ameliorates fatty livers but not obesity or insulin resistance in Lep(ob)/Lep(ob) mice. J Biol Chem 277:19353–19357
Jiang G, Li Z, Liu F et al (2005) Prevention of obesity in mice by antisense oligonucleotide inhibitors of stearoyl-CoA desaturase-1. J Clin Invest 115:1030–1038
Gutierrez-Juarez R, Pocai A, Mulas C et al (2006) Critical role of stearoyl-CoA desaturase-1 (SCD1) in the onset of diet-induced hepatic insulin resistance. J Clin Invest 116:1686–1695
Kakuma T, Lee Y, Unger RH (2002) Effects of leptin, troglitazone, and dietary fat on stearoyl CoA desaturase. Biochem Biophys Res Commun 297:1259–1263
Backhed F, Ding H, Wang T et al (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A 101:15718–15723
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1131
Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006) Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023
Mijalski T, Harder A, Halder T et al (2005) Identification of coexpressed gene clusters in a comparative analysis of transcriptome and proteome in mouse tissues. Proc Natl Acad Sci USA 102:8621–8626
Dumas ME, Wilder SP, Bihoreau MT et al (2007) Direct quantitative trait locus mapping of mammalian metabolic phenotypes in diabetic and normoglycemic rat models. Nat Genet 39:666–672
Dumas ME, Maibaum EC, Teague C et al (2006) Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for large-scale epidemiological research: the INTERMAP Study. Anal Chem 78:2199–2208
Acknowledgements
The authors thank C. Hetherington (Biomedical Services, University of Oxford) for invaluable help with the implementation of the mouse colony. A. A. Toye, M. E. Dumas and C. Blancher contributed equally to this work. This work was funded by the Wellcome Functional Genomics Initiative BAIR (Biological Atlas of Insulin Resistance) (066786). D. Gauguier holds a Wellcome Senior Fellowship in Basic Biomedical Science (057733). S. P. Wilder is a recipient of a Wellcome Prize Studentship in Bioinformatics and Statistical Genetics.
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Effects of fat feeding on gene transcription in the liver of 129S6 mice (PDF 337.9 kb).
Supplementary Table 2
Oligonucleotides designed for quantitative real-time PCR analysis of gene transcription regulation in 129S6 and BALB/c mice (PDF 30.1 kb).
Rights and permissions
About this article
Cite this article
Toye, A.A., Dumas, M.E., Blancher, C. et al. Subtle metabolic and liver gene transcriptional changes underlie diet-induced fatty liver susceptibility in insulin-resistant mice. Diabetologia 50, 1867–1879 (2007). https://doi.org/10.1007/s00125-007-0738-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-007-0738-5