Abstract
Aims/hypothesis
Sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase (SNARK) is involved in cellular stress responses linked to obesity and type 2 diabetes. We determined the role of SNARK in response to metabolic stress and insulin action on glucose and lipid metabolism in skeletal muscle.
Methods
Vastus lateralis skeletal muscle biopsies were obtained from normal glucose tolerant (n = 35) and type 2 diabetic (n = 31) men and women for SNARK expression studies. Primary myotube cultures were derived from biopsies obtained from normal glucose tolerant individuals for metabolic studies.
Results
SNARK (also known as NUAK2) mRNA expression was unaltered between normal glucose tolerant individuals and type 2 diabetic patients. SNARK expression was increased in skeletal muscle from obese (BMI >31 kg/m2) normal glucose tolerant individuals and type 2 diabetic patients (1.4- and 1.4-fold, respectively, p < 0.05) vs overweight (BMI <28 kg/m2) normal glucose tolerant individuals and type 2 diabetic patients. SNARK mRNA was increased in myotubes exposed to palmitate (12-fold; p < 0.01), or TNF-α (25-fold, p < 0.05), but not to oleate, glucose or IL-6, whereas expression of the AMP-activated protein kinase α2 subunit was unaltered. Small interfering (si)RNA against SNARK reduced mRNA and protein in myotubes by 61% and 60%, respectively (p < 0.05). SNARK siRNA was without effect on basal or insulin-stimulated glucose uptake or lipid oxidation, and insufficient to rescue TNF-α- or palmitate-induced insulin resistance.
Conclusions/interpretation
Skeletal muscle SNARK expression is increased in human obesity, and in response to metabolic stressors, but not type 2 diabetes. Partial SNARK depletion failed to modify either glucose or lipid metabolism, or protect against TNF-α- or palmitate-induced insulin resistance in primary human myotubes.
Similar content being viewed by others
Introduction
The ability to adjust energy use and storage in times of fuel surplus or deficiency is vital for survival throughout evolution. AMP-activated protein kinase (AMPK) is known as a master regulator of glucose and lipid metabolism in mammals through sensing of the ratio of AMP:ATP in the cell [1, 2]. Upon activation of AMPK, anabolic energy-demanding pathways are switched off and catabolic ATP-producing pathways are activated, thereby controlling peripheral glucose and lipid homeostasis. Recently, a family of AMPK-related kinases has been described, thereby broadening the regulation of metabolism in response to cellular stress to include novel protein kinases [3].
AMPK is a heterotrimeric complex consisting of a catalytic AMP-sensitive α subunit and regulatory β and γ subunits. Twelve protein kinases (BRSK1, BRSK2, NUAK1, NUAK2, QIK, QSK, SIK, MARK1, MARK2, MARK3, MARK4 and MELK) closely related to AMPKα1 and AMPKα2 have been identified in the human kinome, thus forming a 14 kinase phylogenetic tree known as ‘AMPK-related kinases’ [3, 4]. LKB1, an upstream activating kinase of AMPK, phosphorylates and activates 11 of the AMPK-related kinases [3]. Although the role of AMPK-related kinases is largely unknown, they do not appear to interact with the regulatory β and γ subunits of AMPK [5]. Furthermore, the majority of the AMPK-related kinases are not activated by AMP or pharmacological activators of AMPK including 5-aminoimidazole-4-carboxamide-1β-4-ribofuranoside (AICAR) and phenformin [3, 6]. Recent evidence suggests that skeletal muscle-selective knockout of LKB1 increases insulin sensitivity [7], possibly implicating these novel AMPK-related protein kinases in the regulation of glucose homeostasis.
Haploinsufficient Snark (also known as Nuak2) knockout mice develop obesity, hepatic steatosis, altered serum lipid profiles, hyperinsulinaemia, hyperglycaemia and impaired glucose tolerance with age [8]. SNARK was first identified as an ultraviolet B-induced gene in keratinocytes [9]. SNARK is located on human chromosome 1q32 and is translated into a single protein of approximately 76 kDa [10]. In rats, Snark is expressed in multiple tissues, with highest expression in kidney [11]. Sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase (SNARK) is activated in a cell-type-specific manner by a variety of stressors including hyperosmotic stress, DNA damage and oxidative stress, as well as nutrients including glucose and glutamine [10, 11]. Several aspects of SNARK activation and regulation are broadly similar to AMPK [12]. For example, SNARK and AMPK are both AMP-responsive and activated by treatments known to increase the AMP:ATP ratio, including glucose deprivation and chemical ATP production [10, 11, 13]. Nevertheless, the metabolic role of SNARK, particularly in humans and at the cellular level in skeletal muscle, is incompletely resolved.
Given that Snark-haploinsufficient mice become obese and glucose-intolerant [8], this AMPK-related protein kinase may play a permissive role in energy metabolism. Thus, we tested the hypothesis that SNARK is involved in cellular stress responses associated with obesity and type 2 diabetes. Since skeletal muscle plays an important role in the regulation of whole-body glucose uptake, the aim of this study was to determine the role of SNARK in response to metabolic stress and insulin action on glucose and lipid metabolism in human skeletal muscle.
Methods
Study participants and muscle biopsy procedure
Biopsies were obtained using a Weil–Blakesley conchotome from vastus lateralis skeletal muscle under local anaesthesia (lidocaine hydrochloride 5 mg/ml) from normal glucose tolerant and type 2 diabetic men and women for mRNA expression analysis of the gene encoding SNARK. Biopsies were snap-frozen and stored in liquid nitrogen until use. The clinical characteristics of the study participants are presented in Table 1. HOMA2-insulin resistance (IR) values were determined using the HOMA calculator (www.dtu.ox.ac.uk). The regional ethical committee at Karolinska Institutet approved all study protocols. Informed written and verbal consent was received from all study participants.
Skeletal muscle cell cultures and in vitro treatments
Skeletal muscle cell cultures were established from satellite cells isolated from vastus lateralis skeletal muscle biopsies obtained from normal glucose tolerant individuals. Satellite cells were isolated by trypsin–EDTA digestion, and myoblasts were cultured and differentiated into myotubes as previously described [14]. Myotubes were used to test the effects of a variety of cellular stressors, including nutrients, hormones or cytokines, on SNARK mRNA expression. Myotubes were treated with palmitate (0.25 mmol/l), oleate (0.25 mmol/l), glucose (25 mmol/l), TNF-α (20 ng/ml) or IL-6 (20 ng/ml) for 2 or 7 days. SNARK mRNA expression was determined as described below.
To determine the direct role of SNARK on basal and insulin-stimulated glucose and lipid metabolism, myoblasts were transfected with small interfering (si) RNA against a scrambled non-specific sequence or SNARK (80 pmol) (Ambion/Applied Biosystems, Foster City, USA) for 16 h before initiation of differentiation and also after 2 days into the differentiation programme using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in serum- and antibiotic-free DMEM [15]. Glucose incorporation into glycogen and lipid oxidation were determined, as described below.
SNARK mRNA expression
SNARK mRNA expression was assessed in vastus lateralis skeletal muscle biopsies or primary skeletal muscle cell cultures (myotubes) using quantitative RT-PCR (ABI PRISM 7000 Sequence Detector System; Applied Biosystems). Total RNA was purified from skeletal muscle biopsies using Trizol reagent (Invitrogen, Carlsbad, CA, USA) and from myotubes using an RNeasy Mini Kit (Invitrogen). Purified RNA was treated with DNase I using a DNA-free kit (Ambion) and cDNA synthesis was performed with a SuperScript First Strand Synthesis System (Invitrogen). TaqMan gene expression assays (Hs00388292_m1 and Hs00178903_m1) for SNARK and AMPKα2 (also known as PRKAA2) mRNA expression were independently used in multiplex, with β2-microglobulin as a reference gene (Applied Biosystems). All samples where assayed in duplicate. The mRNA relative abundance was calculated using the standard curve method as described by Applied Biosystems.
Glycogen synthesis in skeletal muscle cells
Glucose incorporation into glycogen was determined as described [16]. Myotubes were incubated in the absence or presence of either 6 or 60 nmol/l insulin for 30 min and thereafter incubated for 90 min with d-[U-14C]glucose (3700 Bq/ml; Amersham, Uppsala, Sweden) in DMEM (1 g glucose/l). When indicated, myotubes were exposed to either palmitate (0.25 mmol/l) or TNF-α (20 ng/ml) for 48 h before the experiment was performed. Experiments were performed in duplicate. Results are reported as nmol glucose (mg protein)−1 h−1.
Lipid oxidation in skeletal muscle cells
NEFA oxidation was assessed by exposing myotubes to [3H]palmitic acid and measuring the production of 3H-labelled water. Myotubes were cultured in six well plates and serum starved overnight. Cells were washed once with PBS and incubated for 5.5 h with 1 ml DMEM (1 g glucose/l), supplemented with fatty acid-free BSA (0.2% wt/vol.) and [9,10(n)-3H]palmitic acid (1850 Bq/ml; Amersham), with or without insulin (120 nmol/l) or AICAR (1 mmol/l; Toronto Research Chemicals, Toronto, ON, Canada). To absorb non-metabolised palmitate, 0.2 ml of the cell supernatant was mixed with 0.8 ml charcoal slurry (0.1 g charcoal powder in 1 ml 0.02 mol/l TRIS–HCl buffer, pH 7.5) in a 2 ml Eppendorf tube and shaken for 30 min. Samples were subjected to centrifugation at 18,500×g for 15 min, after which 0.3 ml supernatant with 3H-labelled-bound water was withdrawn, and radioactivity was determined by liquid scintillation counting (WinSpectral 1414 Liquid Scintillation Counter; Wallac, Turku, Finland). Each experiment was performed in duplicate. Results are reported as per cent converted palmitate (mg protein)−1 h−1.
Western blot analysis
Lysates were prepared from myotubes transfected with siRNA against a scrambled sequence or SNARK. Proteins were separated by SDS-PAGE and subjected to immunoblot analysis with an antibody directed against SNARK (Proteintech, Manchester, UK). Proteins where visualised by enhanced chemiluminescence and quantified by densiometry. Results are reported as arbitrary units and normalised to a loading control (desmin).
Statistics
Results are presented as means ± SE. Differences between groups were determined by Student’s t test or two-way ANOVA. When ANOVA was applied, pair-wise multiple comparison procedures were performed using the Holm–Sidak method at a significance level of 0.05.
Results
Skeletal muscle SNARK mRNA expression is increased with obesity but not in diabetes
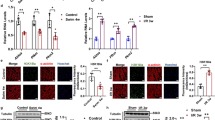
SNARK mRNA expression was determined in vastus lateralis skeletal muscle biopsies from normal glucose tolerant and type 2 diabetic men and women (Fig. 1). Type 2 diabetic patients had fasting hyperglycaemia, and elevated HbA1c levels, compared with normal glucose tolerant individuals (p < 0.05). Individuals were stratified into two groups based on BMI. The metabolic derangements were exacerbated in the obese (BMI >31 kg/m2) vs overweight (BMI <28 kg/m2) type 2 diabetic patients, who also had increased HOMA2-IR values (p < 0.05). SNARK mRNA expression was increased 1.4-fold in obese normal glucose tolerant individuals (BMI >31 kg/m2) vs overweight normal glucose tolerant individuals (BMI <28 kg/m2). SNARK mRNA was also increased 1.4-fold in obese type 2 diabetic patients (BMI >31 kg/m2) vs overweight type 2 diabetic patients (BMI <28 kg/m2). SNARK mRNA expression was similar in normal glucose tolerant and type 2 diabetic patients, irrespectively of BMI.
SNARK mRNA expression was determined in vastus lateralis skeletal muscle biopsies from normal glucose tolerant individuals and type 2 diabetic patients with BMI <28 kg/m2 (white bars) and BMI >31 kg/m2 (black bars). Result are reported in arbitrary units (A.U.) as means ± SE for n = 14–18 individuals. *p < 0.05 compared with normal glucose tolerant individuals with BMI <28 kg/m2
Effects of cellular stressors on SNARK mRNA expression
SNARK mRNA expression was assessed in cultured myotubes treated with a variety of nutrients or cytokines for 2 days (Fig. 2a). Exposure of myotubes to either palmitate (12-fold; p < 0.01) or TNF-α (25-fold, p < 0.05), but not oleate, glucose or IL-6, increased SNARK mRNA expression compared with untreated myotubes, whereas expression of the AMPKα2 subunit was unaltered (Fig. 2b). Similar responses on SNARK mRNA expression were also observed in myotubes exposed to these metabolic stressors for 7 days (data not shown). Furthermore, SNARK mRNA expression was unaltered following exposure of myotubes to AMPK activators, including AICAR, metformin or rosiglitazone (data not shown).
Effect of cellular stressors on SNARK mRNA expression. Cultured myoblasts were treated with palmitate (0.25 mmol/l), oleate (0.25 mmol/l), glucose (25 mmol/l), TNF-α (20 ng/ml) or IL-6 (20 ng/ml) for 2 days followed by assessment of SNARK mRNA expression (a) and AMPKα2 mRNA expression (b). Results are reported in arbitrary units (A.U.) as means ± SE for n = 5 individual cultures. *p < 0.05 compared with unstimulated control cells
SNARK silencing in myotubes
The effect of SNARK gene silencing on mRNA expression (Fig. 3a) and protein level (Fig. 3b) was determined in cultured myotubes. siRNA against SNARK reduced mRNA expression by 61% and protein content by 60% compared with myotubes transfected with siRNA against a scrambled sequence.
SNARK gene silencing in primary human myotubes. The effect of SNARK siRNA was determined on mRNA expression (a) and protein levels (b). Results are reported in arbitrary units (A.U.) as means ± SE for n = 4 individual cultures. *p < 0.05 compared with myotubes treated with siRNA against a scrambled (Scr) sequence. IB, immunoblot
Role of SNARK on glucose incorporation into glycogen
Glucose incorporation into glycogen was assessed in myotubes transfected with siRNA against a scrambled sequence or SNARK (Fig. 4). Insulin increased glucose incorporation into glycogen in myotubes transfected with siRNA against a scrambled sequence (1.8- and 2.3-fold in the presence of 6 or 60 nmol/l insulin, respectively; p < 0.05 vs basal). Similar effects of insulin on glucose incorporation into glycogen were noted for myotubes transfected with siRNA against SNARK (1.8- and 2.4-fold increase in the presence of 6 or 60 nmol/l insulin, respectively; p < 0.05 vs basal). SNARK silencing did not influence basal and insulin-stimulated glucose incorporation into glycogen.
Role of SNARK in glucose incorporation into glycogen. Glucose incorporation into glycogen was measured in myotubes transfected with either siRNA against a scrambled sequence (Scr) or SNARK and incubated in the absence (basal; white bars) or presence of a submaximal (6 nmol/l; grey bars) or maximal (60 nmol/l; black bars) concentration of insulin. Results are means ± SE for n = 8 individuals cultures. *p < 0.05 compared with basal scrambled siRNA-transfected myotubes
Role of SNARK on lipid oxidation
Lipid oxidation was determined in myotubes transfected with siRNA against a scrambled sequence or SNARK (Fig. 5). Insulin suppressed lipid oxidation in myotubes transfected with siRNA against a scrambled sequence (14%; p < 0.05) and this effect was unaltered by SNARK silencing. AICAR increased lipid oxidation in myotubes transfected with siRNA against either a scrambled sequence (3.5-fold; p < 0.01) or SNARK (3.2-fold; p < 0.01) vs the basal condition. Thus, SNARK silencing did not influence basal or insulin- or AICAR-stimulated lipid oxidation.
Role of SNARK in lipid oxidation. Lipid oxidation was measured in myotubes transfected with siRNA against either a scrambled sequence (Scr) or SNARK and incubated in the absence (basal; white bars) or presence of either insulin (60 nmol/l; grey bars) or AICAR (1 mmol/l; black bars). Results are means ± SE for n = 8 individuals cultures. *p < 0.05 compared with basal scrambled siRNA-transfected myotubes
Effect of SNARK silencing on TNF-α- or palmitate-induced insulin resistance
Given that TNF-α and palmitate treatment markedly increased SNARK mRNA expression, we also examined whether protein levels were altered under these conditions (Fig. 6a). Exposure of myotubes to either TNF-α or palmitate for 48 h increased SNARK protein content (p < 0.05). SNARK silencing reduced protein content by 56% (p < 0.05), and completely prevented the effect of either TNF-α or palmitate to increase SNARK protein.
Effect of SNARK silencing on TNF-α- and palmitate-induced insulin resistance. Human myotubes were transfected with either siRNA against a scrambled sequence (Scr) or SNARK and incubated in the absence (basal; white bars) or presence of TNF-α (20 ng/ml; grey bars) or palmitate (0.25 nmol/l; black bars) for 48 h. The effect of SNARK siRNA silencing was determined on protein levels (a), and glucose incorporation into glycogen was measured in the absence or presence of insulin (b). Results are means ± SE for n = 4 individuals. *p < 0.05 compared with myotubes incubated in the absence of insulin. †p < 0.05 compared with myotubes incubated in the absence of TNF-α and palmitate (basal). A.U. arbitrary units; IB, immunoblot
Exposure of myotubes to TNF-α or palmitate induces insulin resistance in cultured human myotubes [17, 18], coincident with increased SNARK expression (Fig. 2a). Thus, we determined whether SNARK silencing prevents insulin resistance arising from these factors (Fig. 6b). Myotubes were transfected with siRNA against either a scrambled sequence or SNARK and incubated for 48 h in the absence or presence of TNF-α (20 ng/ml) or palmitate (0.25 mmol/l). TNF-α exposure decreased insulin-stimulated glucose incorporation into glycogen by 41% in the presence of 6 nmol/l (p < 0.05), but not 60 nmol/l insulin, in myotubes treated with siRNA against either a scrambled sequence or SNARK. Thus, siRNA against SNARK does not protect against TNF-α-induced insulin resistance. Palmitate exposure decreased insulin-stimulated glucose incorporation into glycogen by 37% in the presence of 60 nmol/l insulin (p < 0.01), but not 6 nmol/l insulin, in myotubes treated with siRNA against either a scrambled sequence or SNARK. Thus, siRNA against SNARK does not protect against palmitate-induced insulin resistance.
Discussion
The AMPK-related kinase SNARK plays a role in whole-body glucose and energy homeostasis [8]. Whole-body Snark-haploinsufficient mice are obese and glucose-intolerant, implicating an important role for this AMPK-related protein kinase in the regulation of energy and glucose homeostasis [8]. Here we tested the hypothesis that SNARK is involved in cellular stress responses linked to the development of insulin resistance in obesity and type 2 diabetes. We provide evidence that SNARK mRNA expression is increased in skeletal muscle from obese normal glucose tolerant and type 2 diabetic men and women. The increase in SNARK mRNA was unaltered between normal glucose tolerant and type 2 diabetic individuals irrespectively of BMI, indicating the influence of obesity, rather than diabetes per se. Based on the phenotype of the Snark-haploinsufficient mice [8], an increase in SNARK mRNA expression would be predicted to have a positive effect on energy and glucose homeostasis, but this does not appear to be the case, at least in humans. We also report that elevated levels of TNF-α or palmitate, systemic factors implicated in obesity-induced insulin resistance, increased SNARK expression in cultured human skeletal muscle myotubes. Nevertheless, a 60% reduction in SNARK mRNA by siRNA-mediated gene silencing (an effect similar to that achieved in whole-body Snark-haploinsufficient mice [8]), was without effect on basal or insulin-stimulated glucose uptake and lipid oxidation. Furthermore, SNARK silencing did not prevent TNF-α- or palmitate-induced insulin resistance in skeletal muscle. Thus, while obesity and associated systemic factors, such as TNF-α or palmitate increase SNARK mRNA expression, insulin resistance induced by these factors occurs via a SNARK-independent mechanism.
Snark-haploinsufficient mice, with a 50% reduction of whole-body Snark mRNA expression, develop obesity and insulin resistance, as evidenced by increased adiposity, fatty liver, increased serum triacylglycerol, hyperinsulinaemia, hyperglycaemia and glucose intolerance [8]. Thus, our findings of increased skeletal muscle SNARK mRNA expression in obese humans and in response to elevated levels of TNF-α or palmitate in vitro, suggest that the metabolic phenotype observed in total-body Snark-haploinsufficient mice is unlikely to arise from a direct effect of SNARK in skeletal muscle. Rather, these results implicate a role for SNARK in other tissues, particularly since skeletal muscle expresses relatively low levels of SNARK compared with liver, kidney, intestine and testis in rodents [11]. The development of mature-onset obesity in whole-body Snark-heterozygous mice might occur from a central defect that modifies food intake and activity levels, rather than a direct effect in peripheral tissues. The absence of a basal energetic defect in skeletal muscle of Snark-heterozygous mice, as well as an unexplained increased motivation/drive for voluntary exercise supports this contention [19]. Fibre type composition and number is unaffected in Snark-heterozygous mice, indicating Snark expression and activity in tissues other than skeletal muscle may contribute to the regulation of glucose and energy homeostasis and motivation for physical activity. For example, impairments in brown adipose tissue may contribute to the obesity phenotype, since whole-body temperature and energy expenditure was reduced in these mice. Further studies using tissue-specific Snark knockout mice may resolve this paradox.
The identification of an AMPK subfamily [3] raises questions about the involvement of these protein kinases in whole-body and cellular glucose and energy homeostasis, as well as the possibility that they might regulate cellular and whole-body metabolic and gene regulatory responses in a manner analogous to AMPK. Lkb1 (also known as Stk11) knockout mice have been important in the evaluation of the metabolic role of the AMPK-related kinases, since it is a master kinase that activates the majority of the members of this subfamily [3]. For example, contraction-mediated glucose uptake is inhibited in skeletal muscle from Lkb1 knockout mice [20] and kinase-dead Ampkα2 transgenic mice [21], but not Ampkα1 and Ampkα2 isoform-specific knockout mice [22], indicating both AMPK-dependent and AMPK-independent pathways may play a role. Moreover, skeletal muscle-selective knockout of Lkb1 increases whole-body and skeletal muscle insulin sensitivity and improves glucose homeostasis [7], implicating AMPK-related protein kinases participate in the negative regulation of insulin action. Given our observation that SNARK mRNA expression was elevated in skeletal muscle from obese humans and myotubes exposed to TNF-α or palmitate, we tested the hypothesis that SNARK silencing would directly alter insulin action on glucose and lipid metabolism. However, neither basal nor insulin-stimulated glucose incorporation into glycogen was altered by SNARK silencing in primary human myotubes. Furthermore, basal, as well as insulin- or AICAR-stimulated beta oxidation was unchanged by SNARK silencing. This contrasts with earlier findings which revealed that AMPKα subunit silencing prevents the effects of IL-6 to promote beta oxidation into human myotubes [23]. Thus, AMPK, rather than SNARK, appears to control lipid oxidation in skeletal muscle. Although siRNA against SNARK reduced mRNA expression by 60%, we cannot exclude the possibility that a total knockout of SNARK may be required to reveal effects on glucose and lipid metabolism.
Our observation of increased SNARK mRNA expression in skeletal muscle from obese humans, and in response to TNF-α or palmitate treatment, potentially connects SNARK to metabolic regulation in response to cellular stress signals. For example, TNF-α secretion from adipose tissue is increased and IL-6 serum levels are elevated in obese insulin-resistant individuals [24]. Thus, the intracellular communication between adipocytes or macrophages in obesity may provide stress signals that increase SNARK expression in skeletal muscle, which may modify insulin action. Nevertheless, siRNA against SNARK failed to prevent the development of TNF-α- or palmitate-induced insulin resistance on glucose incorporation into glycogen. Previous studies from our laboratory provide evidence that siRNA-mediated reduction of either the mitogen-activated protein kinase kinase kinase kinase isoform 4 [15] or the inhibitor of nuclear factor-kappaβ (NF-κβ) kinase [17] prevents TNF-α-induced insulin resistance in human skeletal muscle. Interestingly, SNARK has anti-apoptotic properties, acting through a TNF-α-sensitive nuclear NF-κβ-mediated mechanism [25]. Thus, the increase in SNARK expression may be a consequence, rather than a cause of skeletal muscle insulin resistance.
In conclusion, we provide evidence against a role for SNARK in the regulation of skeletal muscle glucose or lipid metabolism. Nevertheless, obesity, but not type 2 diabetes, is associated with an increase in skeletal muscle SNARK mRNA expression in humans. This increase in SNARK mRNA expression may occur as a consequence of systemic factors associated with metabolic impairments in obesity, since exposure of myotubes to elevated levels of TNF-α or palmitate acutely increased SNARK mRNA expression. siRNA against SNARK failed to rescue TNF-α- or palmitate-induced insulin resistance, indicating changes in SNARK expression occur as a consequence, rather than a cause of insulin resistance. Based on our findings in human skeletal muscle, and the insulin-resistant and obesity phenotype in whole-body Snark-haploinsufficient mice [8], SNARK expression in metabolically active tissues beyond skeletal muscle may play a role in whole-body energy and glucose homeostasis.
Abbreviations
- AICAR:
-
5-Aminoimidazole-4-carboxamide-1β-4-ribofuranoside
- AMPK:
-
AMP-activated protein kinase
- IR:
-
Insulin resistance
- siRNA:
-
Small interfering RNA
- SNARK:
-
Sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase
References
Long YC, Zierath JR (2008) Influence of AMP-activated protein kinase and calcineurin on metabolic networks in skeletal muscle. Am J Physiol Endocrinol Metab 295:E545–E552
Hardie DG (2008) AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 32(Suppl 4):S7–S12
Lizcano JM, Goransson O, Toth R et al (2004) LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J 23:833–843
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298:1912–1934
Al-Hakim AK, Goransson O, Deak M et al (2005) 14-3-3 cooperates with LKB1 to regulate the activity and localization of QSK and SIK. J Cell Sci 118:5661–5673
Sakamoto K, Goransson O, Hardie DG, Alessi DR (2004) Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab 287:E310–E317
Koh HJ, Arnolds DE, Fujii N et al (2006) Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol 26:8217–8227
Tsuchihara K, Ogura T, Fujioka R et al (2008) Susceptibility of Snark-deficient mice to azoxymethane-induced colorectal tumorigenesis and the formation of aberrant crypt foci. Cancer Sci 99:677–682
Rosen CF, Poon R, Drucker DJ (1995) UVB radiation-activated genes induced by transcriptional and posttranscriptional mechanisms in rat keratinocytes. Am J Physiol 268:C846–C855
Lefebvre DL, Bai Y, Shahmolky N et al (2001) Identification and characterization of a novel sucrose-non-fermenting protein kinase/AMP-activated protein kinase-related protein kinase, SNARK. Biochem J 355:297–305
Lefebvre DL, Rosen CF (2005) Regulation of SNARK activity in response to cellular stresses. Biochim Biophys Acta 1724:71–85
Egan B, Zierath JR (2009) Hunting for the SNARK in metabolic disease. Am J Physiol Endocrinol Metab 296:E969–E972
Kuga W, Tsuchihara K, Ogura T et al (2008) Nuclear localization of SNARK; its impact on gene expression. Biochem Biophys Res Commun 377:1062–1066
Al-Khalili L, Kramer D, Wretenberg P, Krook A (2004) Human skeletal muscle cell differentiation is associated with changes in myogenic markers and enhanced insulin-mediated MAPK and PKB phosphorylation. Acta Physiol Scand 180:395–403
Bouzakri K, Zierath JR (2007) MAP4K4 gene silencing in human skeletal muscle prevents tumor necrosis factor-alpha-induced insulin resistance. J Biol Chem 282:7783–7789
Al-Khalili L, Chibalin AV, Kannisto K et al (2003) Insulin action in cultured human skeletal muscle cells during differentiation: assessment of cell surface GLUT4 and GLUT1 content. Cell Mol Life Sci 60:991–998
Austin RL, Rune A, Bouzakri K, Zierath JR, Krook A (2008) siRNA-mediated reduction of inhibitor of nuclear factor-kappaB kinase prevents tumor necrosis factor-alpha-induced insulin resistance in human skeletal muscle. Diabetes 57:2066–2073
Sabin MA, Stewart CE, Crowne EC et al (2007) Fatty acid-induced defects in insulin signalling, in myotubes derived from children, are related to ceramide production from palmitate rather than the accumulation of intramyocellular lipid. J Cell Physiol 211:244–252
Ichinoseki-Sekine N, Naito H, Tsuchihara K et al (2009) Provision of a voluntary exercise environment enhances running activity and prevents obesity in Snark-deficient mice. Am J Physiol Endocrinol Metab 296:E1013–E1021
Sakamoto K, McCarthy A, Smith D et al (2005) Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J 24:1810–1820
Mu J, Brozinick JT Jr, Valladares O, Bucan M, Birnbaum MJ (2001) A role for AMP-activated protein kinase in contraction- and hypoxia-regulated glucose transport in skeletal muscle. Mol Cell 7:1085–1094
Jorgensen SB, Viollet B, Andreelli F et al (2004) Knockout of the alpha2 but not alpha1 5′-AMP-activated protein kinase isoform abolishes 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranosidebut not contraction-induced glucose uptake in skeletal muscle. J Biol Chem 279:1070–1079
Al-Khalili L, Bouzakri K, Glund S, Lonnqvist F, Koistinen HA, Krook A (2006) Signaling specificity of interleukin-6 action on glucose and lipid metabolism in skeletal muscle. Mol Endocrinol 20:3364–3375
Kern PA, Ranganathan S, Li C, Wood L, Ranganathan G (2001) Adipose tissue tumor necrosis factor and interleukin-6 expression in human obesity and insulin resistance. Am J Physiol Endocrinol Metab 280:E745–E751
Legembre P, Schickel R, Barnhart BC, Peter ME (2004) Identification of SNF1/AMP kinase-related kinase as an NF-kappaB-regulated anti-apoptotic kinase involved in CD95-induced motility and invasiveness. J Biol Chem 279:46742–46747
Acknowledgements
This study was supported by grants from the European Research Council, the European Foundation for the Study of Diabetes, the Swedish Research Council, the Swedish Diabetes Association, the Strategic Research Foundation, the Knut and Alice Wallenberg Foundation, the Novo Nordisk Research Foundation and the Commission of the European Communities (Contract No LSHM-CT-2004-005272 EXGENESIS and Contract No LSHM-CT-2004-512013 EUGENE2).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rune, A., Osler, M.E., Fritz, T. et al. Regulation of skeletal muscle sucrose, non-fermenting 1/AMP-activated protein kinase-related kinase (SNARK) by metabolic stress and diabetes. Diabetologia 52, 2182–2189 (2009). https://doi.org/10.1007/s00125-009-1465-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00125-009-1465-x