Abstract
Summary
An automated image processing method is presented for simulating areal bone mineral density measures using high-resolution peripheral quantitative computed tomography (HR-pQCT) in the ultra-distal radius. The accuracy of the method is validated against clinical dual X-ray absorptiometry (DXA). This technique represents a useful reference to gauge the utility of novel 3D quantification methods applied to HR-pQCT in multi-center clinical studies and potentially negates the need for separate forearm DXA measurements.
Introduction
Osteoporotic status is primarily assessed by measuring areal bone mineral density (aBMD) using 2D dual X-ray absorptiometry (DXA). However, this technique does not sufficiently explain bone strength and fracture risk. High-resolution peripheral quantitative computed tomography (HR-pQCT) has been introduced as a method to quantify 3D bone microstructure and biomechanics. In this study, an automated method is proposed to simulate aBMD measures from HR-pQCT distal radius images.
Methods
A total of 117 subject scans were retrospectively analyzed from two clinical bone quality studies. The distal radius was imaged by HR-pQCT and DXA on one of two devices (Hologic or Lunar). Areal BMD was calculated by simulation from HR-pQCT images (aBMDsim) and by standard DXA analysis (aBMDdxa).
Results
The reproducibility of the simulation technique was 1.1% (root mean-squared coefficient of variation). HR-pQCT-based aBMDsim correlated strongly to aBMDdxa (Hologic: R 2 = 0.82, Lunar: R 2 = 0.87), though aBMDsim underestimated aBMDdxa for both DXA devices (p < 0.0001). Finally, aBMDsim predicted aBMD at the proximal femur and lumbar spine with equal power compared to aBMDdxa.
Conclusion
The results demonstrate that aBMD can be simulated from HR-pQCT images of the distal radius. This approach has the potential to serve as a surrogate forearm aBMD measure for clinical HR-pQCT studies when axial bone mineral density values are not required.
Similar content being viewed by others
Introduction
Osteoporosis is a condition characterized by a loss of bone mass and deterioration of bone structural integrity resulting in compromised bone strength and an increased risk of fracture [1]. Currently, evaluation of osteoporotic status is primarily based on projectional and volumetric measures of bone mineral density (BMD) using X-ray imaging techniques. While BMD has been shown to have utility in predicting bone strength, it does not entirely determine fracture risk [2, 3] or adequately assess the impact of therapeutic interventions [4, 5]. Accordingly, considerable interest currently exists in the investigation of other factors associated with bone mechanical competence, including whole bone geometry, cortical and trabecular microstructure, and tissue composition. The development and validation of non-invasive, quantitative technologies able to characterize such features is a critical goal for improving the ability to track disease progression and evaluate therapeutic efficacy in clinical research.
Micro-computed tomography has become an important non-invasive tool for investigating a wide range of aspects related to the biology of bone and other calcified tissues [6–8]. Until recently, true 3D assessment of trabecular and cortical bone microstructure has been limited to ex vivo measurements in laboratory microtomography systems [9, 10]. High-resolution peripheral quantitative computed tomography (HR-pQCT) is a promising non-invasive method for in vivo 3D characterization of bone in humans. Similar to traditional quantitative computed tomography (QCT), HR-pQCT provides the ability to quantitatively assess volumetric bone mineral density (vBMD) in a compartmental fashion in the appendicular skeleton (distal radius and tibia). Additionally, it permits quantification of the geometric, microstructural, and biomechanical features of human cortical and trabecular bone [11–13]. As this technology matures, it is important that the utility of new densitometric, structural, and biomechanical endpoints be evaluated in clinically relevant patient populations against standard reference endpoints.
Areal BMD (aBMD), measured by dual X-ray absorptiometry (DXA) is the most widely used surrogate for bone strength, and therefore is an appropriate yardstick for new quantitative techniques based on emerging imaging modalities such as HR-pQCT. In several recent clinical bone quality studies, forearm DXA has been used in compliment to HR-pQCT as a densitometric gold standard, for diagnostic classification, strength prediction, and fracture discrimination [13–18]. However, there are several disadvantages to adding a DXA exam to a clinical HR-pQCT study. These include, but are not limited to, increased logistical complexity, decreased patient retention and compliance, increased cost, and increased radiation dose to the patient. Furthermore, in the context of multi-center studies, the additional burden of cross-site, cross-manufacturer calibration is often necessary [19].
In this study, a method is proposed to simulate DXA-based aBMD measures at the ultra-distal radius using 3D HR-pQCT image data. The algorithm was tested and validated in normative and osteopenic cohorts who underwent HR-pQCT and DXA exams.
Materials and methods
Subjects
HR-pQCT image data from the baseline examinations from two ongoing patient studies were evaluated retrospectively using the aBMD simulation method described below for comparison against aBMD determined by DXA. The first patient cohort is part of a longitudinal investigation into the effects of alendronate on bone microarchitecture and has been described in detail by Kazakia et al. [14]. In short, postmenopausal women (n = 52) defined as osteopenic by WHO criteria [20] were recruited. The women were included if they were between the ages of 45 and 65, and had been postmenopausal for at least one but not more than 6 years. They were required to exhibit low BMD (T-score range −1.1 to −2.5) by DXA either at the lumbar spine or at the total proximal femur, trochanter, or neck regions of interest. The second cohort consisted of a subset of male and female volunteers (female: n = 45, male: n = 20) participating in a normative cross-sectional HR-pQCT study. These subjects spanned a wide range of ages (20 to 78 years) and anthropometrics (BMI range 17 to 39). For this study, DXA screening was not performed prior to enrollment; therefore, no BMD inclusion/exclusion criteria was used.
For both cohorts, history of or evidence for metabolic bone disease other than postmenopausal bone loss was an exclusion criterion, as was treatment within the previous year with any compound known to influence bone turnover. Both study protocols were approved by the UCSF Committee of Human Research, and all subjects gave written informed consent prior to participation.
HR-pQCT
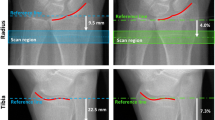
All subjects described below were imaged in a clinical HR-pQCT system (XtremeCT, Scanco Medical AG, Brüttisellen, Switzerland) using the manufacturer’s standard in vivo protocol described in previous patient studies [11, 12, 14]. This system consists of a microfocus X-ray source with a 70-µm focal spot size. The tube voltage was fixed at 60kVp while the current was 900 μA. Filters of 0.3 mm Cu and 1 mm Al are positioned at the aperture to filter soft X-rays in order to reduce patient dose and limit beam-hardening effects. The cone beam X-ray field is incident upon a structured CsI (40 mg/cm2) scintillator coupled by a fiber optic taper to a 2D 3,072 × 256 element CCD detector with a 41-µm pitch. The subject’s forearm was immobilized in a carbon fiber cast that was fixed within the gantry of the scanner. A single dorsal–palmar projection image of the distal radius was acquired to define the tomographic scan region. This region spans 9.02 mm in length (110 slices) and was fixed starting at 9.5 mm proximal from the mid-jointline and extending proximally (Fig. 1a). For tomography, 750 projections were acquired over 180° with a 100-ms integration time at each angular position. The 12.6-cm field of view was reconstructed across a 1,536 × 1,536 matrix using a modified Feldkamp algorithm, yielding 82 µm voxels [21]. Total scan time was 2.8 min with an equivalent dose of approximately 4.2 µSv.
The reconstructed linear attenuation values were converted to hydroxyapatite (HA) mineral densities using a beam-hardening correction and phantom calibration procedure previously described for an ex vivo microtomography system [22]. The calibration phantom (Scanco Medical AG, Brüttisellen, Switzerland) was composed of five cylinders of HA–resin mixtures with a range of mineral concentrations (0, 100, 200, 400, and 800 mg HA/cm3) where 0 mg HA/cm3 represents a soft tissue equivalent background devoid of mineral. The reconstructed images were segmented using semi-automatically drawn contours at the periosteal surface of the radius. The total vBMD of the radius was calculated as the mean calibrated mineral density within this volume of interest (VOI).
Simulation algorithm
Quantitative projection and region of interest (ROI) densitometric analysis of HR-pQCT images was performed using an automated algorithm that aligns and segments the distal forearm in a way that mimics the subject positioning, soft tissue correction, and ROI analysis inherent to clinical DXA examinations of the ultra-distal (UD) radius. The algorithm was developed in C and implemented within the framework of the scanner manufacturer’s Image Processing Language software (IPL v5.06-ucsf, Scanco Medical AG). A flow diagram of the procedure is shown in Fig. 2. The simulated projection images are generated in three primary steps: (1) determination of a common coordinate system, (2) spatial masking of extra-osteal soft tissue, and (3) quantitative projection.
Clinical DXA requires standardized prone positioning of the forearm to ensure reproducible BMD assessment. In contrast, HR-pQCT is acquired with the radius and ulna at a variably oblique angle to the axial coordinate system. It is therefore necessary to define a standard orientation that reflects the patient positioning process inherent to DXA. In order to approximate the DXA scenario, the 3D HR-pQCT images were transformed into a common coordinate system prior to forward projection (Fig. 3). By nature of the patient positioning for HR-pQCT, it was assumed that all datasets approximately share a common z-axis (inferior–superior direction) but have an arbitrary in-plane orientation. The x′-axis was defined as the line shared by the centroids of the radius and ulna for the central slice—corresponding approximately to the anatomical medial–lateral direction. The y′-axis was therefore the third orthogonal axis and approximately corresponds to the dorsal–palmar direction. An in-plane rotational transformation about the midpoint between centroids was applied to bring the voxel coordinate system inline with this common anatomical coordinate system.
The radius and ulna centroids were calculated with respect to the area bound by their respective periosteal surfaces. For the radius, the periosteal surface was defined by a semi-automatically drawn contour generated during the routine HR-pQCT microstructural analysis process [23]. The ulnar periosteal boundary was determined using an automated process (see Fig. 2): First a fixed threshold corresponding to 300 mg HA/cm3 was applied to binarize the grayscale image. The radius was then removed using the contoured VOI described above. A binary dilation (3 pixels) was then performed to close the ulnar cortical shell and component labeling was applied to fill the medullary void space by discarding all but the largest connected void space (corresponding to the background around the ulna). The outer surface was then eroded by 3 pixels to return the ROI boundary approximately to the periosteal edge.
Following alignment in the common coordinate system, the grayscale images were spatially masked using the radius periosteal VOI. In this manner, the ulna and all extra-osseal soft tissue did not contribute to the projected image, approximating the soft tissue compensation inherent to DXA. The masked 3D image was then projected along the dorsal–palmar direction (y′-axis) according to the discrete line integral:
where aBMDsim is the simulated areal bone mineral density of the distal radius projected onto the x′z′-plane (corresponding to medial–lateral and superior–inferior axes), [HA](x′,y′,z′) is the aligned 3D HR-pQCT-calibrated mineral density image matrix, N is the number of voxels in the y′ direction, and Δy is the voxel size in y′. The mean aBMDsim was then calculated as the arithmetic average of all non-zero pixels from this projected image.
Reproducibility
Reproducibility of the aBMDsim measurement was determined in 8 radii of volunteers spanning a large age range (age = 25 to 65 years). Three repeat measurements were performed for each subject with complete repositioning between each scan. For three of the patients, a single dataset was excluded due to excessive motion artifacts visually apparent in the reconstructed images. Therefore, a total of five patients with three scans and three patients with two scans were used to calculate the root mean squared coefficient of variation (RMS-CV%) for aBMDsim.
DXA
Areal bone densitometry data were acquired for the radius, proximal femur, and lumbar spine using one of two commercial DXA scanners; 42 osteopenic women from the first cohort were scanned with the QDR 4500 (Hologic Inc., Bedford, MA, USA) and the remaining 75 subjects were scanned using the Lunar Prodigy (GE Healthcare, Chalfont St. Giles, UK). Standard ROIs used for clinical assessment of osteoporosis status were identified to determine aBMD. The UD region of interest was automatically determined by the scanner software (Fig. 1b, c). For the Hologic device, this region started at the most proximal end of the endplate of the radius and extended 15 mm proximally. For the Lunar device, the region started where the radius and ulna superimpose and extended proximally for 20 mm. Mean BMD values from the UD ROI will subsequently be referred to as aBMDdxa. Areal BMD measures were also determined for the lumbar spine (L1–L4) and total proximal femur using the standard densitometry protocols and analysis software provided by the manufacturer.
Statistics
Mean and standard deviations were calculated for all indices. A paired Student’s t test was used to determine whether aBMDsim measures were statistically different from aBMDdxa. Regression analysis was performed to evaluate how well aBMDsim correlated to aBMDdxa. Previous studies have found differences in absolute BMD measurements between devices from these manufacturers [19, 24]. For this reason, the regression analysis was performed individually for subjects scanned on Lunar and Hologic DXA devices. The regression coefficient of determination values and linear equations relating aBMDsim to aBMDdxa were calculated. In order to evaluate significant differences in the regressions, a two way ANOVA was used with aBMDsim and the device grouping as independent variables. The absolute difference between the simulation and DXA aBMD values was determined and Bland–Altman plots were used to evaluate systematic bias in the simulation assumptions. Lastly, regression analysis was performed between aBMD at the UD radius (simulated and DXA-based) and aBMD for the lumbar spine and total femur.
Results
A representative image of a simulated projection is shown in Fig. 4. The CV% for aBMDsim of the distal radius was determined by repeat acquisitions in eight subjects with complete subject repositioning between scans. The mean aBMDsim of this group was 0.365 ± 0.053 g/cm2 and ranged from 0.269 to 0.431 g/cm2. The RMS-CV% for the eight patients scanned for reproducibility was 1.1%.
The correlation scatter plot and corresponding Bland–Altman plot for aBMDsim against aBMDdxa are shown in Fig. 5. The regression analysis equations are reported in Table 1. There is a clear offset between Hologic and Lunar devices, though aBMDsim correlated strongly to both (Hologic: R 2 = 0.82; Lunar R 2 = 0.87; both p < 0.0001) and significantly underestimated aBMDdxa (p < 0.0001). The underestimation was the result of fixed offsets in the regression equation (Hologic 0.11 g HA/cm2; Lunar 0.04 g HA/cm2; p < 0.0001) while the slopes approached unity for both devices (Hologic 0.94; Lunar 0.91; p = 0.77) with positive intercepts. Compared against either device, aBMDsim was not found to have a strong aBMD dependent trend in the absolute difference between aBMDsim and aBMDdxa (Fig. 5b). Correlation of vBMD determined by HR-pQCT to aBMDdxa was more moderate (R 2 = 0.62 and R 2 = 0.64 for Hologic and Lunar, respectively).
Finally, aBMDdxa of the UD radius and HR-pQCT-derived aBMDsim shared very similar predictive strength for aBMD of the total femur and lumbar spine determined by DXA (Fig. 6). In the Lunar cohort, the correlations were moderately strong for the femur (R 2 = 0.50, p < 0.0001 for both aBMDsim and aBMDdxa) and weak for the spine (R 2 = 0.33 for both aBMDsim and aBMDdxa; p < 0.0001 for both). For the Hologic cohort, which consisted of early postmenopausal subjects with a narrow range of spinal and femoral aBMDdxa, there were no significant correlations to aBMD of the total femur or lumbar spine for either aBMDsim or aBMDdxa at the UD radius (R 2 < 0.02).
Discussion
In this study, we have demonstrated an automated method for simulating areal BMD measures from 3D HR-pQCT images of the ultra-distal radius. Similar techniques have previously been developed for the proximal femur for traditional QCT imaging [25]. This technique would primarily be beneficial for clinical osteoporosis studies as a controlled complement to standard forearm DXA densitometry or where DXA is not available. The algorithm is advantageous in several respects: First, it automatically orients the radius and ulna in a standard anatomic position that approximately corresponds to patient positioning for a clinical DXA examination such that there is no ulnar–radial superposition. In a multi-center, clinical study this would significantly minimize inter-operator variability in patient positioning inherent to DXA. Furthermore, it is reasonable to expect that different HR-pQCT sites have access to DXA devices from different manufacturers. The use of HR-pQCT-derived aBMD measures would avoid variability known to exist between DXA manufacturers [19, 24]. Finally, when appropriate, this approach provides the option of eliminating forearm DXA scans altogether from a clinical research protocol, thereby reducing the minor radiation dose to human subjects subjected to this procedure.
In DXA, two X-ray energies are used to compensate for variable soft tissue attenuation path lengths. In the algorithm presented here, spatial segmentation of the 3D image approximates this compensation by masking peripheral soft tissue and the ulna prior to forward projection. This method does not account for intra-medullary soft tissue (i.e., bone marrow) nor potential compositional variability of the marrow itself (hematopoietic vs. fatty marrow). However, for the ultra-distal radius, these effects are expected to be minimal compared to differences in extra-osseal soft tissue across subjects and compared to axial skeletal sites.
In this study, we have validated the simulation technique against standard clinical DXA of the UD radius in a total of 117 subjects, spanning a large range of ages and BMD values. The algorithm successfully generated projections for all subjects in the study. Reproducibility for measuring aBMDsim (including patient positioning and acquisition) was approximately 1.1% RMS-CV. This is similar to previously reported reproducibility results for standard volumetric BMD indices determined by HR-pQCT [11, 14]. Regression analysis revealed strong correlations (R 2 > 0.8) between aBMDsim and aBMDdxa in the UD radius for a broad patient cohort consisting of male and female volunteers between 20 and 80 years of age (scanned on a GE Lunar device) and a narrow cohort of osteopenic women within 6 years of menopause (scanned on a Hologic device). These results are consistent with correlation coefficients (R 2 = 0.5–0.94) determined for the cross-manufacturer forearm DXA standardization effort commissioned by the International Committee of Standards in Bone Measurement [19] as well as with the results reported for similar algorithms developed for QCT [25]. True vBMD was less well correlated to aBMDdxa. This is not surprising given the size dependence inherent to projectional BMD measures. It follows that simulation of the projection process does significantly improve prediction of DXA-based BMD values.
It is important to note that the standard VOI for a clinical HR-pQCT acquisition (9.02 mm in length) is shorter than the standard ultra-distal ROI prescribed by DXA manufacturers (20 and 15 mm in length for Lunar and Hologic, respectively). Furthermore, each manufacturer uses different anatomical landmarks to localize the ROI. These two facts may partly explain the discrepancy in the coefficients of determination for aBMDsim compared to Lunar and Hologic (R 2 = 0.87 vs. R 2 = 0.82) and the difference in the regression intercept (0.04 vs. 0.11 g/cm2). As expected, the aBMDsim better predicted Lunar aBMDdxa values, where the ROI is more similar with respect to the longitudinal placement compared to the Hologic ROI. The difference in the correlation coefficients also likely reflects the relative variability in the patient cohorts scanned on either device.
As expected, aBMDsim and aBMDdxa of the UD radius were poor to moderate predictors of aBMD at axial skeletal sites (lumbar spine and proximal femur). Despite the significantly smaller analysis ROI, aBMDsim had an equivalent degree of predictive power for DXA aBMD in the lumbar spine and proximal femur. The magnitude of the predictive power for the Lunar cohort was similar to previous studies comparing intersite BMD relations [26, 27]. This group spanned a larger age and BMD range, compared to the Hologic cohort, which was comprised exclusively of osteopenic women with a narrow range of aBMD values at axial skeletal sites.
An important limitation is that this simulation technique is limited to anatomical sites that may be imaged by HR-pQCT. In this study, we have applied the technique to the distal radius, as this is a routine site for clinical densitometry and a common site of osteoporotic fracture (Colles’ fracture). This technique could also be applied to the distal tibia, which is routinely imaged during clinical HR-pQCT exams, and of interest as a load-bearing site. On the other hand, the proximal femur and lumbar spine—critical sites of osteoporotic fracture—are not accessible by HR-pQCT. Furthermore, classification by T or Z scores is not possible for aBMDsim as comparable normative databases are not currently available for HR-pQCT. Therefore, if a clinical study requires densitometry of the axial skeleton or diagnostic classification of osteoporotic status, standard densitometry would be required.
In conclusion, this study has demonstrated that areal BMD of the UD radius can be accurately simulated from 3D HR-pQCT images of the distal radius. This approach has the potential to serve as a surrogate forearm BMD measure for clinical HR-pQCT studies.
References
(1991) Consensus development conference: prophylaxis and treatment of osteoporosis. Am J Med 90:107-110.
Beck TJ, Oreskovic TL, Stone KL, Ruff CB, Ensrud K, Nevitt MC, Genant HK, Cummings SR (2001) Structural adaptation to changing skeletal load in the progression toward hip fragility: the study of osteoporotic fractures. J Bone Miner Res 16:1108–1119
Stone KL, Seeley DG, Lui LY, Cauley JA, Ensrud K, Browner WS, Nevitt MC, Cummings SR (2003) BMD at multiple sites and risk of fracture of multiple types: long-term results from the Study of Osteoporotic Fractures. J Bone Miner Res 18:1947–1954
Black DM, Thompson DE (1999) The effect of alendronate therapy on osteoporotic fracture in the vertebral fracture arm of the Fracture Intervention Trial. Int J Clin Pract Suppl 101:46–50
Delmas PD, Seeman E (2004) Changes in bone mineral density explain little of the reduction in vertebral or nonvertebral fracture risk with anti-resorptive therapy. Bone 34:599–604
Hildebrand T, Laib A, Muller R, Dequeker J, Ruegsegger P (1999) Direct three-dimensional morphometric analysis of human cancellous bone: microstructural data from spine, femur, iliac crest, and calcaneus. J Bone Miner Res 14:1167–1174
Muller R, Ruegsegger P (1997) Micro-tomographic imaging for the nondestructive evaluation of trabecular bone architecture. Stud Health Technol Inform 40:61–79
Ulrich D, van Rietbergen B, Laib A, Ruegsegger P (1999) The ability of three-dimensional structural indices to reflect mechanical aspects of trabecular bone. Bone 25:55–60
David V, Laroche N, Boudignon B, Lafage-Proust MH, Alexandre C, Ruegsegger P, Vico L (2003) Noninvasive in vivo monitoring of bone architecture alterations in hindlimb-unloaded female rats using novel three-dimensional microcomputed tomography. J Bone Miner Res 18:1622–1631
Gasser JA, Ingold P, Grosios K, Laib A, Hammerle S, Koller B (2005) Noninvasive monitoring of changes in structural cancellous bone parameters with a novel prototype micro-CT. J Bone Miner Metab 23:90–96 Suppl
Boutroy S, Bouxsein ML, Munoz F, Delmas PD (2005) In vivo assessment of trabecular bone microarchitecture by high-resolution peripheral quantitative computed tomography. J Clin Endocrinol Metab 90:6508–6515
Khosla S, Riggs BL, Atkinson EJ, Oberg AL, McDaniel LJ, Holets M, Peterson JM, Melton LJ 3rd (2006) Effects of sex and age on bone microstructure at the ultradistal radius: a population-based noninvasive in vivo assessment. J Bone Miner Res 21:124–131
Macneil JA, Boyd SK (2007) Accuracy of high-resolution peripheral quantitative computed tomography for measurement of bone quality. Med Eng Phys 29(10):1096–1105
Kazakia GJ, Hyun B, Burghardt AJ, Krug R, Newitt DC, de Papp AE, Link TM, Majumdar S (2008) In vivo determination of bone structure in postmenopausal women: a comparison of HR-pQCT and high-field MR imaging. J Bone Miner Res 23:463–474
Chavassieux P, Asser Karsdal M, Segovia-Silvestre T, Neutzsky-Wulff AV, Chapurlat R, Boivin G, Delmas PD (2008) Mechanisms of the anabolic effects of teriparatide on bone: insight from the treatment of a patient with pycnodysostosis. J Bone Miner Res 23:1076–1083
Boutroy S, Van Rietbergen B, Sornay-Rendu E, Munoz F, Bouxsein ML, Delmas PD (2008) Finite element analysis based on in vivo HR-pQCT images of the distal radius is associated with wrist fracture in postmenopausal women. J Bone Miner Res 23:392–399
Sornay-Rendu E, Boutroy S, Munoz F, Delmas PD (2007) Alterations of cortical and trabecular architecture are associated with fractures in postmenopausal women, partially independent of decreased BMD measured by DXA: the OFELY study. J Bone Miner Res 22:425–433
Melton LJ 3rd, Riggs BL, van Lenthe GH, Achenbach SJ, Muller R, Bouxsein ML, Amin S, Atkinson EJ, Khosla S (2007) Contribution of in vivo structural measurements and load/strength ratios to the determination of forearm fracture risk in postmenopausal women. J Bone Miner Res 22:1442–1448
Shepherd JA, Cheng XG, Lu Y, Njeh C, Toschke J, Engelke K, Grigorian M, Genant HK (2002) Universal standardization of forearm bone densitometry. J Bone Miner Res 17:734–745
(1994) Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group. World Health Organ Tech Rep Ser 843:1-129
Feldkamp LA, Davis LC, Kress JW (1984) Practical cone-beam algorithm. J Opt Soc Am A 1:612–619
Burghardt AJ, Kazakia GJ, Laib A, Majumdar S (2008) Quantitative assessment of bone tissue mineralization with polychromatic micro-computed tomography. Calcif Tissue Int 83:129–138
Laib A, Hauselmann HJ, Ruegsegger P (1998) In vivo high resolution 3D-QCT of the human forearm. Technol Health Care 6:329–337
Prevrhal S, Lu Y, Genant HK, Toschke JO, Shepherd JA (2005) Towards standardization of dual X-ray absorptiometry (DXA) at the forearm: a common region of interest (ROI) improves the comparability among DXA devices. Calcif Tissue Int 76:348–354
Khoo BC, Brown K, Cann C, Zhu K, Henzell S, Low V, Gustafsson S, Price RI, Prince RL (2008) Comparison of QCT-derived and DXA-derived areal bone mineral density and T scores. Osteoporos Int doi:10.1007/00198-008-0820-y
Augat P, Fuerst T, Genant HK (1998) Quantitative bone mineral assessment at the forearm: a review. Osteoporos Int 8:299–310
Nieves JW, Cosman F, Mars C, Lindsay R (1992) Comparative assessment of bone mineral density of the forearm using single photon and dual X-ray absorptiometry. Calcif Tissue Int 51:352–355
Acknowledgments
The authors would like to thank Dr. Andres Laib and Scanco Medical AG for providing software development support and to acknowledge Thelma Munoz, Jingyi Yu, Nicole Cheng, Melissa Guan, and Ayako Suzuki for their contributions to clinical coordination, DXA and HR-pQCT imaging, and database management. They would also like to thank Dr. Sven Prevrhal of UCSF for helpful technical discussions. This publication was supported by NIH/NCRR UCSF-CTSI grant number UL1 RR024131-01 (AJB), NIH RO1 AG17762 (SM), and NIH F32 AR053446 (GJK). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Conflicts of interest
None.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Burghardt, A.J., Kazakia, G.J., Link, T.M. et al. Automated simulation of areal bone mineral density assessment in the distal radius from high-resolution peripheral quantitative computed tomography. Osteoporos Int 20, 2017–2024 (2009). https://doi.org/10.1007/s00198-009-0907-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00198-009-0907-0