Abstract
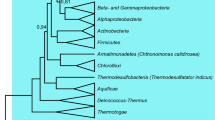
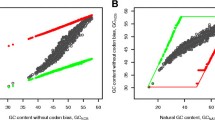
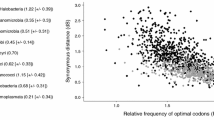
The factors governing codon and amino acid usages in the predicted protein-coding sequences of Tropheryma whipplei TW08/27 and Twist genomes have been analyzed. Multivariate analysis identifies the replicational-transcriptional selection coupled with DNA strand-specific asymmetric mutational bias as a major driving force behind the significant interstrand variations in synonymous codon usage patterns in T. whipplei genes, while a residual intrastrand synonymous codon bias is imparted by a selection force operating at the level of translation. The strand-specific mutational pressure has little influence on the amino acid usage, for which the mean hydropathy level and aromaticity are the major sources of variation, both having nearly equal impact. In spite of the intracellular lifestyle, the amino acid usage in highly expressed gene products of T. whipplei follows the cost-minimization hypothesis. The products of the highly expressed genes of these relatively A + T-rich actinobacteria prefer to use the residues encoded by GC-rich codons, probably due to greater conservation of a GC-rich ancestral state in the highly expressed genes, as suggested by the lower values of the rate of nonsynonymous divergences between orthologous sequences of highly expressed genes from the two strains of T. whipplei. Both the genomes under study are characterized by the presence of two distinct groups of membrane-associated genes, products of which exhibit significant differences in primary and potential secondary structures as well as in the propensity of protein disorder.




Similar content being viewed by others
References
Akashi H, Gojobori T (2002) Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci USA 99:3695–3700
Andersson SG, Sharp PM (1996) Codon usage and base composition in Rickettsia prowazekii. J Mol Evol 42:525–536
Bentley SD, Maiwald M, Murphy LD, et al. (2003) Sequencing and analysis of the genome of the Whipple’s disease bacterium Tropheryma whipplei. Lancet 361:637–644
Berg OG, Silva PJ (1997) Codon bias in Escherichia coli: the influence of codon context on mutation and selection. Nucleic Acids Res 25:1397–1404
Casjens S (1998) The diverse and dynamic structure of bacterial genomes. Annu Rev Genet 32:339–377
Chanda I, Pan A, Dutta C (2005) Proteome composition in Plasmodium falciparum: higher usage of GC-rich nonsynonymous codons in highly expressed genes. J Mol Evol 61:513–523
Das S, Paul S, Chatterjee S, Dutta C (2005a) Codon and amino acid usage in two major human pathogens of genus Bartonella—Optimization between replication-transcriptional selection, translational control and cost minimization. DNA Res 12:91–102
Das S, Pan A, Paul S, Dutta C (2005b) Comparative analyses of codon and amino acid usage in symbiotic island and core genome in nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum. J Biomol Struct Dyn 23:221–232
Das S, Ghosh S, Pan A, Dutta C (2005c) Compositional variation in bacterial genes and proteins with potential expression level. FEBS Lett 579:5205–5210
de Miranda AB, Alvarez–Valin F, Jabbari K, Degrave WM, Bernardi G (2000) Gene expression, amino acid conservation, and hydrophobicity are the main factors shaping codon preferences in Mycobacterium tuberculosis and Mycobacterium leprae. J Mol Evol 50:45–55
dos Reis M, Savva R, Wernisch L (2004) Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res 32:5036–5044
Dufton MJ (1997) Genetic code synonym quotas and amino acid complexity: Cutting the cost of proteins? J Theor Biol 187:165–173
Dutly F, Altwegg M (2001) Whipple’s disease and “Tropheryma whippelii.” Clin Microbiol Rev 14:561–583
Eisen JA, Heidelberg JF, White O, Salzberg SL (2000) Evidence for symmetric chromosomal inversions around the replication origin in bacteria. Genome Biol 1:research 0011.1–0011.9
Fink AL (2005) Natively unfolded proteins. Curr Opin Struct Biol 15:35–41
Foster PG, Jermiin LS, Hickey DA (1997) Nucleotide composition bias affects amino acid content in proteins coded by animal mitochondria. J Mol Evol 44:282–288
Frank AC, Lobry JR (2000) Oriloc: prediction of replication boundaries in unannotated bacterial chromosomes. Bioinformatics 16:560–561
Fuxreiter M, Simon I, Friedrich P, Tompa P. (2004) Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol 338:1015–1026
Garnier J, Gibrat JF, Robson B (1996) GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol 266:540–553
Herbeck JT, Wall DP, Wernegreen JJ (2003) Gene expression level influences amino acid usage, but not codon usage, in the tsetse fly endosymbiont Wigglesworthia. Microbiology 149:2585–2596
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Lafay B, Lloyd AT, McLean MJ, Devine KM, Sharp PM, Wolfe KH (1999) Proteome composition and codon usage in spirochaetes: species-specific and DNA strand-specific mutational biases. Nucleic Acids Res 27:1642–1649
Lafay B, Atherton JC, Sharp PM (2000) Absence of translationally selected synonymous codon usage bias in Helicobacter pylori. Microbiology 146:851–860
Linding R, Russell RB, Neduva V, Gibson TJ (2003) GlobPlot: exploring protein sequences for globularity and disorder. Nucleic Acids Res 31:3701–3708
Lobry JR, Gautier C (1994) Hydrophobicity, expressivity and aromaticity are the major trends of amino-acid usage in 999 Escherichia coli chromosome-encoded genes. Nucleic Acids Res 22:3174–3180
Lopez P, Philippe H (2001) Composition strand asymmetries in prokaryotic genomes: mutational bias and biased gene orientation. CR Acad Sci III 324:201–208
Mackiewicz P, Mackiewicz D, Kowalczuk M, Cebrat S (2001) Flip-flop around the origin and terminus of replication in prokaryotic genomes. Genome Biol 2:interactions 1004.1–1004.4
McInerney JO (1998) Replicational and transcriptional selection on codon usage in Borrelia burgdorferi. Proc Natl Acad Sci USA 95:10698–10703
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nishio Y, Nakamura Y, Kawarabayasi Y, Usuda Y, Kimura E, Sugimoto S, Matsui K, Yamagishi A, Kikuchi H, Ikeo K, Gojobori T (2003) Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res 13:1572–1579
Ohama T, Muto A, Osawa S (1990) Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC- content. Nucleic Acids Res 18:1565–1569
Ohkubo S, Muto A, Kawauchi Y, Yamao F, Osawa S (1987) The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet 210:314–322
Pan A, Dutta C, Das J (1998) Codon usage in highly expressed genes of Haemophillus influenzae and Mycobacterium tuberculosis: translational selection versus mutational bias. Gene 215:405–413
Pascal G, Medigue C, Danchin A (2005) Universal biases in protein composition of model prokaryotes. Proteins 60:27–35
Peixoto L, Fernandez V, Musto H (2004) The effect of expression levels on codon usage in Plasmodium falciparum. Parasitology 128:245–251
Raoult D, Ogata H, Audic S, Robert C, Suhre K, Drancourt M, Claverie JM (2003) Tropheryma whipplei Twist: a human pathogenic Actinobacteria with a reduced genome. Genome Res 13:1800–1809
Ratnaike RN (2000) Whipple’s disease. Postgrad Med J 76:760–766
Ring S, Schneider T, Marth T (2003) Mucosal immune response to Tropheryma whipplei. Int J Med Microbiol 293:69–76
Rispe C, Delmotte F, van Ham RC, Moya A (2004) Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res 14:44–53
Rocha EP (2004) Codon usage bias from tRNA’s point of view: redundancy, specialization, and efficient decoding for translation optimization. Genome Res 14:2279–2286
Rocha EP, Danchin A (2003) Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet 34:377–378
Romero H, Zavala A, Musto H (2000) Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces. Nucleic Acids Res 28:2084–2090
Schaber J, Rispe C, Wernegreen J, Buness A, Delmotte F, Silva FJ, Moya A (2005) Gene expression levels influence amino acid usage and evolutionary rates in endosymbiotic bacteria. Gene 352:109–117
Seligmann H (2003) Cost-minimization of amino acid usage. J Mol Evol 56:151–161
Sharp PM (1991) Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol 33:23–33
Sharp PM, Li WH (1987a) The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res 15:1281–1295
Sharp PM, Li WH (1987b) The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol 4:222–230
Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE (2005) Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res 33:1141–1153
Singer GA, Hickey DA (2003) Thermophilic prokaryotes have characteristic patterns of codon usage, amino acid composition and nucleotide content. Gene 317:39–47
Slowik A, Szczudlik A (2002) Whipple’s disease—a rare cause of neurological symptoms and disorders. Neurol Neurochir Pol 36:959–970
Wright F, Bibb MJ (1992) Codon usage in the G + C–rich Streptomyces genome. Gene 113:55–65
Wu M, Sun LV, Vamathevan J, et al. (2004) Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2:0327–0341
Zavala A, Naya H, Romero H, Musto H (2002) Trends in codon and amino acid usage in Thermotoga maritima. J Mol Evol 54:563–568
Acknowledgments
This work was supported by the Council of Scientific and Industrial Research (Project CMM 0017) and Department of Biotechnology, Government of India (Grant BT/BI/04/055-2001).
Author information
Authors and Affiliations
Corresponding author
Additional information
[Reviewing Editor: Dr. Richard Kliman]
Rights and permissions
About this article
Cite this article
Das, S., Paul, S. & Dutta, C. Evolutionary Constraints on Codon and Amino Acid Usage in Two Strains of Human Pathogenic Actinobacteria Tropheryma whipplei . J Mol Evol 62, 645–658 (2006). https://doi.org/10.1007/s00239-005-0164-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-005-0164-6