Abstract
Ubiquitin (Ub)-conjugating enzymes (E2) are key enzymes in ubiquitination or Ub-like modifications of proteins. We searched for all proteins belonging to the E2 enzyme super-family in seven species (Homo sapiens, Mus musculus, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe, Saccharomyces cerevisiae, and Arabidopsis thaliana) to identify families and to reconstruct each family’s phylogeny. Our phylogenetic analysis of 207 genes led us to define 17 E2 families, with 37 E2 genes, in the human genome. The subdivision of E2 into four classes did not correspond to the phylogenetic tree. The sequence signature HPN (histidine–proline–asparagine), followed by a tryptophan residue at 16 (up to 29) amino acids, was highly conserved. When present, the active cysteine was found 7 to 8 amino acids from the C-terminal end of HPN. The secondary structures were characterized by a canonical alpha/beta fold. Only family 10 deviated from the common organization because the proteins were devoid of enzymatic activity. Family 7 had an insertion between beta strands 1 and 2; families 3, 5 and 14 had an insertion between the active cysteine and the conserved tryptophan. The three-dimensional data of these proteins highlight a strong structural conservation of the core domain. Our analysis shows that the primitive eukaryote ancestor possessed a diversified set of E2 enzymes, thus emphasizing the importance of the Ub pathway. This comprehensive overview of E2 enzymes emphasizes the diversity and evolution of this superfamily and helps clarify the nomenclature and true orthologies. A better understanding of the functions of these enzymes is necessary to decipher several human diseases.
Similar content being viewed by others
Introduction
Ubiquitination is characterized by a rapid and reversible posttranslational covalent fixation of ubiquitin (Ub) onto proteins (Ciechanover 2006). This mechanism, which is part of the specific protein-degradation pathway by the 26S proteasome, plays an important role in targeting proteins as well as intracellular signalling. It has an impact on many cellular functions, such as DNA repair, transcription, signal transduction, endocytosis, and sorting (Welchman et al. 2005).
The ubiquitination of proteins requires three types of enzymes. First, Ub-activating enzymes E1 form a thioester bond with Ub in an adenosine triphosphate–dependent reaction. Second, Ub-conjugating enzymes E2 (or Ubc) carry Ub and transfer it either directly to the substrate protein or to a third type of enzyme, the Ub ligases E3. These latter enzymes facilitate the ligation of Ub to the target protein. The possible intervention of E4 enzymes has been reported in the formation of polyubiquitin chains of lengths >4 Ub (Koegl et al. 1999). Although polyubiquitination generally signals a protein for degradation by the proteasome, monoubiquitination is mainly involved in signaling itself. Deubiquitination, in contrast, is performed by several deubiquitinating enzymes (Wilkinson 1997).
Ub is a small protein of 76 amino acids and highly conserved in eukaryotes. Ub binds to the target protein by way of an isopeptide link between the C-terminal glycine and a lysine residue of the protein, and it appears that three conserved lysines within Ub are crucial for the formation of polyubiquitin chains. Several proteins similar to Ub (Ub-like [Ubl])—such as SUMO1-2-3, NEDD8-RUB1, ATG8, ATG12-APG12, ISG15, FAU, or URM1—can also be used in similar mechanisms. Proteins tagged by SUMO (small Ub modifier) or NEDD8 are not recognized for degradation; however, they play a role in gene transcription activation, protein localization, and stabilization. Each target–function combination has its own unique combination of E1, E2, and E3 enzymes.
The human genome comprises eight genes encoding E1 Ub and Ubl activating enzymes (Ensembl release 50, July 2008) and four genes encoding the two subunits of the E1 enzyme involved in sumoylation (Supplementary Table 1). E3 enzymes are the most numerous, with probably >1000 members (Pickart 2004). They are the most specific in targeting the proteins for ubiquitination and can be divided into two main groups: RING and HECT (homologous to the E6-AP carboxyl terminus) proteins. However, the catalytic modules of these two families are unrelated in sequence or structure. HECT domain-containing E3 enzymes form intermediate thioesters with Ub at their active site cysteine before transferring Ub to substrates, whereas most RING finger domain-containing E3 enzymes act as scaffolds that bind to E2 enzymes and substrates simultaneously (Özkan et al. 2005).
The general signature motif of the E2 enzyme superfamily is an HPN tripeptide (histidine–proline–asparagine) and an active cysteine residue generally located at the eighth amino acid on the C-terminal side of this canonical motif (Cottee et al. 2006) (Fig. 1). In addition, several domains are highly conserved and may play an important role in the function of these enzymes. For example, the sequence between the H2 helix and L3 loop seems to be the binding site for free or ligated Ub (Winn et al. 2004), whereas a highly conserved N-terminal sequence in the core domain may play a role in the interaction with E1 enzymes. To ascertain a high specificity, Ub binds to the target protein by way of a complex mechanism. Because the small number of E1 enzymes cannot allow for great specificity, the selection of the target protein has to come from the other enzymes. However, little is known about the relation between E2 and E3 enzymes. Although several E2 enzymes can interact with the same E3 enzyme, the converse is true as well: one unique E2 enzyme can work with several E3 enzymes as well RING and HECT enzymes. Abundant structural data exist about the interaction of E2 and E3 proteins, implicating the L4 and L7 loops as key sites of E3 interactions (Winn et al. 2004). However, although certain amino acid positions are clearly identified to play a role as sites for E3 binding, other elements on the E2 surface are required to define the specificity of the interaction of any given E2–E3 pair, such as polar contacts involving side chains in H1 helix of the E2 (Pickart 2001). Even if the phenylalanine in position 63 in the L4 loop is essential for HECT interaction and the tryptophan in position 95 in the L7 loop is necessary for RING interaction, it is the overall three-dimensional (3D) surface and charge that significantly contribute to the specificity of interaction with E3 enzymes (Martinez-Noel et al. 2001). In the same way, different E3 enzymes might have slightly different binding surfaces on a same E2 (Özkan et al. 2005). Moreover, supplementary levels of regulation and specificity are mediated by the intervention of additional proteins.
Explanative structure showing in their 3D context the different residues mentioned in Fig. 6. Loops are labeled L1 to L8; β-strands are labeled S1 to S4; and helices are labeled H1 to H4 (with “h” for the generally conserved 3/10 helix). (Adapted from 3D structure of UBE2D2, PDB source E2SK, and legended from Winn et al. 2004)
According to the nature of the E2 enzyme, the type of ubiquitination can be different as well. For example, in yeast, only UBC13 type E2 enzymes are able to form polyubiquitin chains bound on Lys63, and the presence of a Ub-conjugating enzyme variant (UEV) is necessary for this activity (Andersen et al. 2005).
The mechanism of action of the Ubl-conjugating enzymes is thought to be identical to the Ub-conjugating enzymes, with E2 enzymes specific of each Ubl (for example, UBC8 for ISG15, UBC9 for SUMO, UBC12 for NEDD8 in S. cerevisiae). However, these Ubl generally form monoconjugates.
Defining the correct orthologs in a large family of proteins is a difficult task. This is particularly true for the E2 superfamily. In humans, the E2 enzyme superfamily has been estimated to be composed of 33 genes (Lorick et al. 2005). Jiang and Beaudet (2004) have suggested the involvement of as many as 50 genes by including UEV, which lack the critical cysteine residue in the catalytic site. Different numbers of genes are estimated to make up the many E2 enzyme families of various species. For example, 13 genes were identified in Saccharomyces cerevisiae (Jones et al. 2001), 22 in Caenorhabditis elegans (Kipreos 2005), 25 in Drosophila melanogaster (Jones et al. 2001), and 37 in Arabidopsis thaliana (Kraft et al. 2005). In addition, the nomenclature of E2 enzymes varies between species, with E2 enzymes in humans being named UBE2 followed by a letter, whereas in S. cerevisiae they are referenced UBC followed by a number. The numbering of E2 orthologs does not always match between species either.
Several classifications of E2 families have been published (Burroughs et al. 2008; Jones et al. 2001), but no clear consensus has emerged. General trees of the E2 enzyme superfamily have been previously published (Jones et al. 2001; Winn et al. 2004; Melner et al. 2006; Burroughs et al. 2008). However, recent progress in genome sequencing and annotation currently allows for a more comprehensive approach. Although a comprehensive phylogenetic analysis of the E2 enzyme superfamily exists for the D. melanogaster, C. elegans and A. thaliana genomes (Jones et al. 2001; Kipreos 2005; Kraft et al. 2005), no exhaustive analysis has been published for the following species with full genome sequences: Homo sapiens, Mus musculus, Schizosaccharomyces pombe, and S. cerevisiae. Recently, Burroughs et al. (2008) proposed a general tree of various family members of the E2 superfamily, highlighting their particular structures. However, this study did not analyze the “classical Ubc families” in detail, contained several pseudogenes and redundant sequences, and included very distantly related genes.
To identify the orthologs in this family, we made a careful analysis of several complete genomes judiciously distributed in the tree of life. In the present work, we examined seven species with known complete genome sequences to identify E2 enzyme families and to construct the phylogeny of each family using several phylogenetic methods, with the aim to propose a definition for the E2 enzyme families according to the reconstructed orthologies. Finally, we validated our results on the recent release of the full genome of the sea anemone (Nematostella vectensis) (Putnam et al. 2007). Because dysfunction of the ubiquitination pathway may play an important role in several diseases, it is important to identify the true orthologies of E2 enzymes in model organisms and humans.
Materials and Methods
Identification of E2 Proteins
The list of E2 protein sequences from C. elegans and A. thaliana was used as an initial set (Jones et al. 2001; Kipreos 2005; Kraft et al. 2005). Homologs were identified in a first step in the other species based on best hits by BLASTP search (blastp program with default parameters) (http://blast.ncbi.nlm.nih.gov/Blast.cgi), with a cutoff score of 10−20. To identify the next set of E2 enzyme homologs, we used sequences obtained in the first step as new queries and ran another BLASTP search using the National Center for Biotechnology (NCBI) server in the genomes of the seven species: H. sapiens, M. musculus, D. melanogaster, C. elegans, S. pombe, S. cerevisiae and A. thaliana. As queries, we used approximately 150 amino acids of the core domain of the proteins. We only kept RefSeq entries containing NP_ in the accession numbers because these proteins were curated manually with experimental support. Redundant sequences and noninformative pseudogenes were eliminated. Each protein sequence was linked to a gene by submission to GenBank on the NCBI website (http://www.ncbi.nlm.nih.gov/Genbank/).
Generation of Sequence Alignments and Definition of E2 Enzyme Families
We aligned all of the full-length protein sequences obtained to identify the E2 core domains of each sequence. Sequences of the core domains were assembled into a single file using the BioEdit multiple sequence editor (Hall 1999). Multiple protein sequence alignment of truncated sequences was performed using ClustalW algorithm incorporating default settings, and the alignment was refined by manual readjustment. Using the core domains, we also generated a specific human tree of all E2 enzyme sequences to obtain easier visualization of the different families. Several proteins were reallocated to families using their similarity score obtained by the NCBI protein–protein BLAST program. Sequence homology was estimated by the PRSS (Perfect Recognition Similarity Scores) program, which computes the statistical significance of the similarity between two sequences (http://fasta.bioch.virginia.edu/fasta_www2/fasta_www.cgi?rm=shuffle). Proteins were attributed to a family if the PRSS score was <10−30. After using this approach to assign each protein to a group, we realigned the sequences and reanalyzed the relations within each group using full-length amino acid sequence data.
Phylogenetic Analysis
Aligned sequences were first used to generate matrices of distances between proteins based on the Jones–Taylor–Thornton matrix model, and these matrices were used to generate phylogenetic trees according to the minimum evolution (neighbor-joining [NJ]) algorithm using the Phylip3.65 software package (http://evolution.genetics.washington.edu/phylip.html). Bootstrapping of 1000 replicates was performed according to the method of Felsenstein, whose parameters were set on default, with the addition of an outgroup (an A. thaliana gene for the family trees). Phylogenetic trees were visualized and manipulated using TreeView1.6.6 (http://www.treeview.net/) and TreeDyn198.3 (http://www.treedyn.org/) (Chevenet et al. 2006).
Phylogenetic analyses were performed with the maximum likelihood (ML) method using the Proml program of the Phylip3.65 package, a maximum parsimony (MP) method using the Protpars program of the Phylip3.65 package, and a bayesian inference (BI) method using the MrBayes program (http://mrbayes.scs.fsu.edu/). MP and ML methods were used with default parameters. ML calculations were based on the Jones–Taylor–Thornton substitution matrix. Bootstrap support was estimated using 1000 nonparametric replicates for all three methods. For the BI phylogenesis, two simultaneous independent Markov chains were run under Jones’ fixed rate model. To compute the family trees, generations were run until the split frequency score was <0.01 by sampling every 10 generations and with a burn-in of 25% of the number of generations. Each phylogenetic algorithm run was replicated once using another bootstrapped set of data to insure convergence of results.
Construction of Phylogenetic Trees
For each algorithm, a consensus tree of the bootstrap results was obtained using the Consense program of the Phylip3.65 package with the majority rule extended-type option. For the BI tree, numbers indicate the clade credibility values, and branches <95% were collapsed. For the other trees, bootstrap values are indicated; branches carrying bootstrap values under a defined threshold (59% for NJ and ML trees and 85% for MP tree) were collapsed. A consensus tree of the four trees obtained with the different algorithms was generated after inspection of the concordance between the various results and using the Consense program of Phylip3.65 package with default parameters and the majority rule extended-type option. Every tree was displayed and annotated with TreeDyn198.3. Only internodes with significant support in at least three of the analyses were drawn.
Phylogeny of Concatenated Sequences
We selected one ortholog gene from each family in each species. Protein sequences were concatenated in the same order to obtain one sequence per species. This concatenation was used to build a phylogenetic tree of the studied species. The four algorithms were used (NJ, ML, MP, and BI), and the consensus tree was drawn.
Results and Discussion
Inventory of the E2 Enzymes in Seven Species
Our primary goal was to propose a list and classification of the complete set of E2 proteins encoded by the human genome. To obtain a clearer view of the relation and the evolution of this superfamily of proteins, we added several other species with fully sequenced genomes distributed in the tree of life. As the other mammal, we choose the mouse because many transgenic animal studies allow functional evaluations of proteins in this species. C. elegans and D. melanogaster are two multicellular organisms representative of distantly related lineages with many available functional genomic data. All of these species are members of Bilateria in the Animalia phylum. Two distantly related yeast species were chosen to evaluate the ancestral set of E2 proteins in eukaryotes, using information from another phylum (Fungi). Finally, we used genes from A. thaliana as the outgroup to design the phylogenetic trees. Prokaryotic homologs of the E2 enzymes have recently been described in bacteria (Iyer et al. 2006); however, we did not include these too distantly related genes in our study.
We chose to work with proteins rather than nucleotide sequences because mutational noise is less important in amino acid sequences (Inagaki and Roger 2006). Indeed, the fast evolution of nucleotides in the third position of the codons, allowed by the degeneration of the genetic code, produces an accumulation of mutational bias (Jeffroy et al. 2006).
Genbank, Swiss-Prot, TrEMBL (including Pfam and the InterPro database of protein families), Gender and Ensembl were initially used to retrieve a total of 78 RefSeq protein coding sequences in A. thaliana, 16 in S. cerevisiae, 15 in S. pombe, 31 in C. elegans, 36 in D. melanogaster, 181 in M. musculus, and 71 in H. sapiens.
We eliminated redundant sequences and pseudogenes as well as sequences for which PRSS scores were <1. This led us to exclude the TSG101-UEVLD family, the UFC1 family, and the Ub conjugation–like ATG3 and ATG10 enzymes. These families may be considered distantly related or converged to similar 3D structures.
Our final list includes 48 E2 protein sequences in A. thaliana, 14 in S. cerevisiae, 14 in S. pombe, 26 in C. elegans, 32 in D. melanogaster, 36 in M. musculus, and 37 in H. sapiens. Table 1 and Supplementary Table 2 list all 207 E2 proteins, including their main characteristics, species of origin, chromosomal localization, synonyms, name and length of the corresponding deduced protein, identified homologues, and GenBank accession numbers.
General Features of the E2 Enzyme Families in the Seven Species
In a first step, to identify the main groups of proteins (families) with maximum confidence, we aligned truncated protein sequences to avoid long-branch attractions and to minimize noise from C- and N-terminal extremities. The definition of the central core was arbitrary in its details; however, we verified that small differences in the definition of the core sequences had no influence on the obtained results (data not shown). In contrast, the large set of studied sequences (n = 207) gave a good idea of the general organization of the primary structures of these proteins. Compared with pair-wise alignments, multiple ones allowed for better definitions of orthologous sequences. It was suggested that for distant species, a minimum of 20 sequences needed to be aligned to obtain good results (Margulies et al. 2006). The study of several distantly related species facilitated the recognition of the minimum relations inside the families (the core signatures).
We defined the limits of the superfamily by fixing the PRSS score of protein sequences to be <10−30. The alignment of all protein sequences, the global phylogenetic analysis, and the computing of similarity scores showed the existence of 17 subgroups (see Supplementary Figs. 1, 2, and 3), which we named “families.” Others have classified E2 enzyme proteins into 18 groups by splitting family 3 into three groups (XIII, XIV, and XV) and family 4 into two groups (IV and V) while overseeing families 16 and 17 (Jones et al. 2001).
Usual classes of E2 enzymes were defined by the presence or absence of an N and/or C extension. The most frequent class was class 1, containing only the core domain. Among the 17 families, 5 contained >1 class, suggesting that the notion of class generally has no phylogenetic meaning.
A large part of the gene diversity in the different species can be represented by the 14 genes of S. cerevisiae. We used the known nomenclature of yeast, or Caenorhabditis genes, to classify the families; however, more functional information is necessary to propose a better nomenclature.
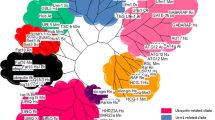
All families had at least one member in humans (Fig. 2). Chromosomal locations of each E2 coding gene in the human genome are drawn on the karyotype representation in Supplementary Fig. 4. Figure 3 depicts the distribution of the genes in each family in the 7 species. It is possible to distinguish 4 types of E2 enzyme families, taking into account their species distribution. Ten families are present in all species (families 1 to 10); 2 families are present in all species except C. elegans (families 11 and 12); 4 families are only absent in the 2 yeasts (families 13 to 16); and 1 family is present only in Bilateria (family 17).
The phylogeny of a family of proteins is important to identify the true homology of proteins (orthology) among different species. Using this information, it is then possible to create a 3D structural model of the candidate proteins and/or to assign biologic functions to them. Studying a large protein superfamily through various species is extremely difficult; therefore, it is no surprise that we found several errors in the ortholog nomenclature in the literature (Supplementary Table 3).
As mentioned previously, the primary sequence signature HPN, followed by a tryptophan residue at 16 (up to 29) amino acids, is highly conserved. This tryptophan 95 has not been shown to make contact with the HECT or the RING domain. However, the crystal structure analysis of the complexes with E2 and either RING or HECT E3 proteins reveals that the side chain of Trp95 is positioned closely to Pro97 at the tip of loop L7 as well as to Pro65 and Pro66 at the base of loop L4 (Martinez-Noel et al. 2001). Pro65 and Pro66 are found in a motif strongly conserved (Y/FPxxPP) 7 to 11 amino acids from the N-terminal side of the HPN motif. Interaction between Trp95 and the proline residues might stabilize the L7 loop and contribute to the correct positioning of the L4 and L7 loops relative to each other. Ala98 of the L7 loop seems to be important for interaction because it makes direct contact with the HECT or the RING domain; however, it is not necessary and is not conserved (Martinez-Noel et al. 2001).
When present, the active cysteine is found at seven to eight amino acids from the C-terminal side of HPN. The analysis of primary and secondary structures highlights several original features of certain families (see specific results in later text).
3D Structure and Protein Organization
A complete analysis of all known sequences based on comparative modeling is available elsewhere (Winn et al. 2005) and on the following Web site: http://www.ubiquitin-resource.org. All known experimental X-ray diffraction crystal 3D structures of E2 enzymes are listed in Supplementary Table 4. Fifteen of 17 families have at least 1 member with known experimental 3D structure, and schematic structures are depicted in Supplementary Fig. 5. The available 3D data of the proteins highlight strong conservation of the structure of the core domain. This information may help to rapidly classify new genes and assign specific functions, such as substrate specificity.
Phylogenetic Analysis for the Classification of E2 Proteins
We used the four main classical algorithms for phylogenetic reconstruction, and the results were mainly coherent. Because one of the four methods gave different results in several cases, we only kept the results of the three convergent methods. The construction of consensus trees allowed for simple and meaningful representation of theses results. However, although only the nodes of the trees were considered meaningful; the time scale or relative evolution speeds of branches were lost with such an approach.
In the next paragraphs, we provide short descriptions and indicate the main characteristics for each family (Fig. 4 and Supplementary Fig. 6), adding some information on known functions, although a complete review on this subject is beyond the scope of the present work. Depending on the cases, the order of the families was chosen according to the numbering order in S. cerevisiae or C. elegans. Family 10 is an exception and was placed at this position because it belongs to the 10 families with members in all species studied.
Examples of phylogenetic trees for “simple” families, such as family 7 (a), and complex families, such as families 3 (b) and 4 (c). Each tree represents the consensus of four algorithms (NJ, ML, MP, and BI). Only branches present in at least three algorithms are shown, whereas others are collapsed. Numbers indicate the number of algorithms supporting the presence of the node
The first six families can be considered “classical E2 enzyme” families. The hallmark of family 1 was an important C-terminal ubiquitin-associated (UBA) supplementary domain linked to the core domain by a flexible tether of approximately 20 amino acids. This UBA domain is important for polyubiquitination by allowing binding to a second subunit of Ub. The MP analysis identified ubc-20_Ce as the closest C. elegans gene to mammals and Drosophila, suggesting that this gene was the ortholog of the other genes. In contrast, ubc-21_Ce, ubc-22_Ce, and ubc-23_Ce, diverged and can be considered paralogs.
Family 2 had a classical structure without particularities; however, it can be observed that in both mammals we found two genes, suggesting a duplication of the UBC2 gene in their common ancestor, whereas mouse had an additional third gene.
Families 3 and 4 are the only families that possessed two members in both yeasts S. cerevisiae and S. pombe. Family 3 was characterized by two specific regions. One single amino acid insertion altered the orientation of the turn between the first two β-strands in the UBC7_Sc crystallized protein (glutamate 31 in sequence PKSENNIF); however, this glutamate was not conserved in the other members of the family. An insertion of 13 extra residues followed the conserved motif of the active site (HxPGDDPxxxExx) and corresponded to the 3/10 “h” helix. We also identified three subfamilies, each containing different human genes: UBE2G1 (Watanabe et al. 1996), UBE2G2 (Katsanis and Fisher 1998), and two UBE2R (Plon et al. 1993) genes. We obtained different results with the MP analysis compared with other methods, so this tree was excluded in the consensus tree of family 3.
With its 40 members, family 4 was the largest E2 enzyme family, and some members of this family were the most difficult to assign to a particular subfamily. The proline of the HPN signature of the superfamily was not conserved in family 4 and was replaced by a cysteine.
Family 5 missed the canonical tripeptide HPN, which was replaced by TPNGRF or TANGRF. This observation led to the characterization of this family under the NCUBE denomination (non canonical ubiquitin conjugating enzymes) (Lester et al. 2000). There was an insertion of two amino acids (aspartate-aromatic) between strand 4 and helix 2 on the C-terminal side of the active cysteine, whose structure was unknown. The orientation of helix 3 and 4 was nonclassical, with this C-terminal extremity corresponding to a hydrophobic transmembrane domain for association with the endoplasmic reticulum. These enzymes had electrostatic potentials that were more similar to the small Ub modifier (SUMO)–conjugating family 7 orthologs (Winn et al. 2007). For this family we obtained discordant results with species of known phylogenetics, although analyses were recomputed several times, changing the order of input sequences and bootstrap values.
In family 6, we found a duplication of the ancestral gene in D. melanogaster and probably two duplications of the ancestral gene in A. thaliana. All other species possessed only one ortholog gene.
Families 7 to 9 are particular because of the conjugation of Ubl. The proteins in family 7 conjugated SUMO. Like family 3, there were two insertions—one of five residues (positions 32 to 37) between β-strands 1 and 2 and one of two residues near the active cysteine (glu-asp at position –2 and –3 from the conserved tryptophan—rather than asp-lys (Tong et al. 1997)). The N-terminal helix had a nonclassical electrostatic surface, which may be involved in the recognition of SUMO (Giraud et al. 1998). This surface, similar to family 5, was involved in ubiquitination (Winn et al. 2007). We found a duplication of the ancestral gene in C. elegans; however, one of these genes diverged greatly and appeared near the root (MP analysis).
The proteins of family 8 conjugated NEDD8-RUB1. Family 8 had a specific N-terminal extension of 26 residues involved in neddylation (VanDemark and Hill 2004), which was not shown on the 3D model (Supplementary Fig. 5). Like family 5, this family was difficult to analyze because of duplication of the mammalian genes, which seemed to have diverged greatly, creating an apparent subfamily. However, the four mammalian genes appeared at the place most distant from the root in the tree.
The proteins of family 9 conjugated Ub and ISG15. Family 9 had an N-terminal extension not shown in Fig. 4, which may have been involved in recognition of ISG15.
The particularity of family 10 is that no protein had an active cysteine; therefore, they were named “variants of Ubc” or UEV. However, this nomenclature was also used for other proteins that did not belong to this family, such as UEVLD and TSG101. Proteins of family 10 were devoid of enzymatic activity and had no canonical HPN motif. The two last alpha helices were missing.
In families 11 and 12, all species possessed only 1 ortholog gene, except C. elegans, which had no identifiable member, whereas A. thaliana possessed 2 homologs. Family 12 possessed a specific N-terminal supplementary domain (not shown in Supplementary Fig. 5). For this family, analyses were run several times; however, each time we obtained unexpected results. This was likely caused by the fact that the D. melanogaster gene was anchored near the root and that the genes of yeast and mammals diverged. This suggests that the Drosophila gene evolved rapidly because this lineage separated. A Blast search for homologs of these two families lost in C. elegans was run in the 31 whole-sequenced species of nematodes using NemaBLAST (http://www.nematode.net/BLAST/). No member was found except for family 11, for which UBE2S_Hs seemed to have high homology with the XI04817 gene from Xiphinema index (e value 8.8e−51). This species belonged to an early clade of the Nematode group (Blaxter et al. 1998), which would indicate that the genes of these 2 families were lost successively during the evolution of Nematodes.
The loss of four families in yeast does not appear to be phylum specific because genes of families 13 to 16 were present in other species belonging to the Fungi phylum (Candida albicans, Yarrowia lipolytica, Aspergillus terreus, A. fumigatus, A. nidulans, A. oryzae, Coccidioides immitis, Neurospora crassa, Gibberella zeae, Magnaporthe grisea, and Chaetomium globosum). This raises the question of the general relevance of the two yeasts as models in the study of E2 enzyme mechanisms. The missing gene families may be replaced on a functional level by genes obtained from duplications in other families because no organism that we analyzed had <14 E2 enzyme genes.
Families 13 to 17 possessed no member in both yeast species S. cerevisiae and S. pombe. In family 13, in the absence of an asparagine residue in the tripeptide HPH, the enzymes of this family should have had no catalytic activity (Wu et al. 2003). A supplementary domain was present at the N-terminus (RLQKEL and GAPGTLYxyE, x = A or E and y = G or N).
Family 14 had no available 3D structure. There was an insertion of seven amino acids between the active cysteine and the conserved tryptophan, containing the conserved sequence TWxG and corresponding to a small “h” helix. These enzymes had an N-terminal extension.
Family 15 had no evident structural particularity. Theses enzymes were known to conjugate Ub and ISG15 (Zhao et al. 2004).
Family 16 had no known 3D structure, and we found no evident consensus primary sequence. Only one A. thaliana and the C. elegans orthologs had an active cysteine. RCE1 from A. thaliana was a RUB1-NEDD8–conjugating enzyme.
Family 17 lacked any member in both yeasts S. cerevisiae and S. pombe and in A. thaliana. This family had a particular orientation of helix 4 and 3. Strangely, there was no HPN motif in any but the human UBE2Q2 sequence. There is a large extension at the N-terminal extremity not shown in Fig. 4. Family 17 is present only in Bilateria and probably evolved from one of the initial ancestral genes; however, the phylogenetic information was lost in our set of species. This family was the only clade-specific family that we were able to identify and may have participated in the evolution of the Bilateria lineage. Further analysis of other species may narrow the precise period of the apparition of this family.
The 10 families present in all species may correspond to the minimal number of initial genes in the ancestors of eukaryotes. However, it is more probable that the common ancestor of all 3 phyla already possessed a set of 18 ancestral genes in 16 families, given the fact that A. thaliana possessed genes of 16 families. C. elegans lost 2 families (family 11 and 12), and yeasts lost 4 families (families 13 to 16). The genome of A. thaliana was the richest in E2 enzyme genes, indicating the importance of this pathway in plants. Several events of genome duplications were at the origin of this rich set of UBC genes in the lineage of Arabidopsis (Adams and Wendel 2005). A schematic representation of this discussion is proposed in Fig. 5.
The general tree of all proteins (Supplementary Fig. 3) illustrates that we cannot describe the relations between the different families with high confidence. Although more information could be gained by adding species from several other clades, it is also possible that the phylogenetic information contained in the primary sequences has been lost once and for all because of the long evolution of E2 genes in the common ancestors of all eukaryotes. Identification of specific primary sequence signatures or spatial signatures may possibly aid in distinguishing subgroups of families, such as the “ICLDIL” subgroup (see Fig. 6 for a summary of hallmarks of all families).
Schematic of primary and secondary structures summarizing the hallmarks of the 17 E2 enzyme families. Alpha helices are represented by rectangles; β-strands are represented by arrows; and several consensus sequences are highlighted. UBA = UBA domain at the C-terminus in family 1. Family-specific insertions are illustrated by loops. The PxxPP sequence, the active cysteine, and the conserved tryptophan are boxed
Although our definition of families was pragmatic, a biologic significance of such a classification can be determined. The timescale, however, was clearly not the significance. First, the subdivisions inside families 3 and 4 were anterior to the separation between the Animalia and Fungi phyla, a separation estimated to have taken place 1.3 billion years ago (Feng and Doolittle 1997). Second, the unique family specific to Bilateria organisms represented a late “invention” because the separation of Bilateria from other organisms has been estimated to have occurred 615 million years ago (Peterson et al. 2004). Therefore, the proposed classification more probably represents strong selective functional pressures rather than real evolution time.
A functional significance that we could expect from a protein classification would be information about interactions with the protein partners evolving in parallel, such as the E3 enzymes. Unfortunately, available sequence information does not allow the drawing of a general picture. In fact, nothing is known about E3 interaction for families 13, 16, and 17. Only families 3, 4, 8, 11, and 15 are known to interact with an HECT E3, and no clear common sequence signature emerges in these families. Allosteric communication between the E3-binding and E2 active site relies on a complex structural unit formed by a large network of coevolving residues instead of a linear pathway consisting of a small set of residues (Özkan et al. 2005).
The topology of the family trees was generally coherent with known phylogenetic data; however, the family 5 tree was quite complex. It may be proposed that a duplication of the ancestral gene occurred in Bilateria and that one of these duplicated genes (ubc-26_Ce) may have strongly diverged in C. elegans. Ubc-26_Ce probably belonged to subfamily UBEJ2_Hs because subfamily UBE2J1_Hs already included two homolog genes, and subfamily UBE2J2_Hs was devoid of any homolog. Ubc-6_Ce and ubc-15_Ce were probably duplications in the C. elegans lineage in the UBEJ2_Hs subfamily, whereas the UBE2J1_Hs ortholog of Drosophila must have been lost because the nearest gene in Drosophila was CG5823_Dm, which was an ortholog of UBE2J2_Hs (e value 6.e−31 by way of blastp).
UEV proteins are as old as E2 proteins (Villalobo et al. 2002). Family 10 is a good example because it contains several UEV proteins that have been highly conserved in eukaryotes, from Protists to Humans (Andersen et al. 2005). Other families also contain several other UEV proteins; e.g., uev-3_Ce belongs to family 4, but uev-2_Ce is not an ortholog of the human UEV2-UBE2V2. The human UEV3-UEVLD and TSG101 proteins belong to another distant family and therefore were not included in our study, exemplifying that UEV proteins are a polyphyletic heterogeneous group.
Phylogeny of the Species
We found only 10 ortholog genes present in all 7 species (listed in Table 2). These 10 well-defined sequences were concatenated and used to build a phylogenetic tree (Supplementary Fig. 7). Although the use of many genes in concatenation does not guarantee a tree representative of the true historic evolution (Jeffroy et al. 2006), our analysis of this concatenated set allowed for a clear definition of the clades obtained by molecular and morphologic approaches. However, phylogenetic information of this subset of genes was not sufficient to distinguish the precise branching order of Caenorhabditis and Drosophila, underlining the difficulty of this analysis (Blair et al. 2002; Dopazo and Dopazo 2005). Five families have a D. melanogaster gene closer to the mammals’ gene than the C. elegans member, whereas only 3 families have a C. elegans gene closer to the mammals’ genes. Taking a simple majority rule, this is in accordance with the most recent results placing insects nearest to mammals (Wolf et al. 2004). Our results also confirmed that the majority of C. elegans genes evolved more rapidly than their Drosophila counterparts (Mushegian et al. 1998).
Application of our Classification on the Nematostella Genome
There are 46 sequences on the Web site of the Nematostella genome that are identified as Ubc proteins (http://genome.jgi-psf.org/cgi-bin/ToGo?accession=GO:0004840&species=Nemve1&model=1&batchId=34). Among them, 4 sequences (Nemve1:1191, Nemve1:2031, Nemve1:2043, and Nemve1:2091) have PRSS scores too low to be considered true Ubc members. Furthermore, no known Ubc motif is detectable by inspection in the sequence of these proteins. Using Human or S. cerevisiae core sequences as queries, a Blast search yielded 43 Nematostella sequences, the previous 42 “true” genes and a supplementary sequence (Nemve1:152221), which probably corresponds to a pseudogene (C-terminal end of the protein) or a partially identified gene homologous to Nemve1:158438. Inspection of the alignment of these protein sequences with the 207 sequences of our previous set confirmed that most of these sequences matched perfectly the consensus sequences of the defined families. Several families contained genes that evolved more rapidly and that were less clearly characterized (for example Nemve1:169077 belongs to family 5 but lacks the active cysteine, or Nemve1:85975 belongs to family 7 but has an insertion of 3 amino acids before the active site). This analysis confirms the usefulness of our classification and the richness of the genome of this deeply rooted multicellular organism.
Conclusion
This work attempts to clarify the nomenclature and the orthologies of E2 proteins. We focused on seven well-known species with sequenced whole genomes. This analysis highlights the particularities of each species, which are important when searching for functions and orthologs in animal models. While in this study, we found that two families were lost in C. elegans and four families in the yeast species, we also “discovered” a family specific of the kingdom Animalia. The classification we propose should serve as an initial platform, and requires the analysis of additional species to obtain a general view of the E2 enzymes in eukaryotes. Further investigations are warranted and will need to focus on searching for E2 enzyme partners, including the interacting E3 proteins, and defining the precise substrates of each E1-E2-E3 enzyme association (Ub or Ubl and proteins). By defining the role of the Ub-conjugating enzymes in human, it will become possible to understand their implication in various diseases. Abnormal production or regulation of some E2 enzymes has been increasingly connected to diseases of the central nervous system as well as to cancer development. Improving the understanding of the E2 enzyme families in normal and pathologic situations could lead to the development of novel drugs targeting specific E2 enzymes.
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Andersen P, Zhou H, Pastushok L, Moraes T, McKenna S, Ziola B, Ellison M, Dixit V, Xiao W (2005) Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. J Cell Biol 170:745–755
Blair JE, Ike K, Gojobori T, Hedges SB (2002) The evolutionary position of nematodes. BMC Evol Biol 2:7
Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, Vanfleteren JR, Mackey LY, Dorris M, Frisse LM et al (1998) A molecular evolutionary framework for the phylum Nematoda. Nature 392:71–75
Burroughs AM, Jaffee M, Iyer LM, Aravind L (2008) Anatomy of the E2 ligase fold: Implications for enzymology and evolution of ubiquitin/Ub-like protein conjugation. J Struct Biol 162:205–218
Chevenet F, Brun C, Bañuls A, Jacq B, Christen R (2006) TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinform 7:439
Ciechanover A (2006) The ubiquitin proteolytic system: From a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology 66(Suppl):S7–S19
Cottee PA, Abs El-osta YG, Nisbet AJ, Gasser RB (2006) Ubiquitin-conjugating enzyme genes in Oesophagostomum dentatum. Parasitol Res 99:119–125
Dopazo H, Dopazo J (2005) Genome-scale evidence of the nematode-arthropod clade. Genome Biol 6:R41
Feng DF, Doolittle RF (1997) Converting amino acid alignment scores into measures of evolutionary time: a simulation study of various relationships. J Mol Evol 44:361–370
Giraud MF, Desterro JM, Naismith JH (1998) Structure of ubiquitin-conjugating enzyme 9 displays significant differences with other ubiquitin-conjugating enzymes which may reflect its specificity for sumo rather than ubiquitin. Acta Crystallogr D Biol Crystallogr 54:891–898
Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Inagaki Y, Roger AJ (2006) Phylogenetic estimation under codon models can be biased by codon usage heterogeneity. Mol Phylogenet Evol 40:428–434
Iyer LM, Burroughs AM, Aravind L (2006) The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol 7:R60
Jeffroy O, Brinkmann H, Delsuc F, Philippe H (2006) Phylogenomics: The beginning of incongruence? Trends Genet 22:225–231
Jiang YH, Beaudet AL (2004) Human disorders of ubiquitination and proteasomal degradation. Curr Opin Pediatr 16:419–426
Jones D, Crowe E, Stevens TA, Candido EP (2001) Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Biol 3:research0002.1–research0002.15 [epub]
Katsanis N, Fisher EM (1998) Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae ubc7 gene. Genomics 51:128–131
Kipreos ET (2005) C. elegans cell cycles: Invariance and stem cell divisions. Nat Rev Mol Cell Biol 6:766–776
Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S (1999) A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644
Kraft E, Stone S, Ma L, Su N, Gao Y, Lau O, Deng X, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139:1597–1611
Lester D, Farquharson C, Russell G, Houston B (2000) Identification of a family of noncanonical ubiquitin-conjugating enzymes structurally related to yeast UBC6. Biochem Biophys Res Commun 269:474–480
Lorick KL, Jensen JP, Weissman AM (2005) Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol 398:54–68
Margulies EH, Chen CW, Green ED (2006) Differences between pair-wise and multi-sequence alignment methods affect vertebrate genome comparisons. Trends Genet 22:187–193
Martinez-Noel G, Müller U, Harbers K (2001) Identification of molecular determinants required for interaction of ubiquitin-conjugating enzymes and RING finger proteins. Eur J Biochem 268:5912–5919
Melner MH, Haas AL, Klein JM, Brash AR, Boeglin WE, Nagdas SK, Winfrey VP, Olson GE (2006) Demonstration of ubiquitin thiolester formation of UBE2Q2 (UBCi), a novel ubiquitin-conjugating enzyme with implantation site-specific expression. Biol Reprod 75:395–406
Mushegian AR, Garey JR, Martin J, Liu LX (1998) Large-scale taxonomic profiling of eukaryotic model organisms: A comparison of orthologous proteins encoded by the human, fly, nematode, and yeast genomes. Genome Res 8:590–598
Özkan E, Yu H, Deisenhofer J (2005) Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci USA 102:18890–18895
Peterson KJ, Lyons JB, Nowak KS, Takacs CM, Wargo MJ, McPeek MA (2004) Estimating metazoan divergence times with a molecular clock. Proc Natl Acad Sci USA 101:6536–6541
Pickart CM (2001) Mechanisms underlying ubiquitination [review]. Annu Rev Biochem 70:503–533
Pickart CM (2004) Back to the future with ubiquitin. Cell 116:181–190
Plon SE, Leppig KA, Do HN, Groudine M (1993) Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci USA 90:10484–10488
Putnam NH, Srivastava M, Hellsten U, Dirks B, Chapman J, Salamov A, Terry A, Shapiro H, Lindquist E, Kapitonov VV et al (2007) Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317(5834):86–94
Tong H, Hateboer G, Perrakis A, Bernards R, Sixma TK (1997) Crystal structure of murine/human Ubc9 provides insight into the variability of the ubiquitin-conjugating system. J Biol Chem 272:21381–21387
VanDemark AP, Hill CP (2004) Grabbing E2 by the tail. Nat Struct Mol Biol 11:908–909
Villalobo E, Morin L, Moch C, Lescasse R, Hanna M, Xiao W, Baroin-Tourancheau A (2002) A homologue of CROC-1 in a ciliated protist (Sterkiella histriomuscorum) testifies to the ancient origin of the ubiquitin-conjugating enzyme variant family. Mol Biol Evol 19:39–48
Watanabe TK, Kawai A, Fujiwara T, Maekawa H, Hirai Y, Nakamura Y, Takahashi E (1996) Molecular cloning of UBE2G, encoding a human skeletal muscle-specific ubiquitin-conjugating enzyme homologous to UBC7 of C. elegans. Cytogenet Cell Genet 74:146–148
Welchman R, Gordon C, Mayer J (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6:599–609
Wilkinson K (1997) Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J 11:1245–1256
Winn PJ, Religa TL, Battey JN, Banerjee A, Wade RC (2004) Determinants of functionality in the ubiquitin conjugating enzyme family. Structure 12:1563–1574
Winn PJ, Battey JN, Schleinkofer K, Banerjee A, Wade RC (2005) Issues in high-throughput comparative modelling: A case study using the ubiquitin E2 conjugating enzymes. Proteins 58:367–375
Winn PJ, Zahran M, Battey JN, Zhou Y, Wade RC, Banerjee A (2007) Structural and electrostatic properties of ubiquitination and related pathways. Front Biosci 12:3419–3430
Wolf YI, Rogozin IB, Koonin EV (2004) Coelomata and not Ecdysozoa: Evidence from genome-wide phylogenetic analysis. Genome Res 14:29–36
Wu PY, Hanlon M, Eddins M, Tsui C, Rogers RS, Jensen JP, Matunis MJ, Weissman AM, Wolberger C, Pickart CM (2003) A conserved catalytic residue in the ubiquitin-conjugating enzyme family. EMBO J 22:5241–5250
Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yan W, Shulman BA, Huibregtse JM, Krug RM (2004) The ubcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-α/β-induced ubiquitin-like protein. PNAS 101:7578–7582
Acknowledgments
We thank Thierry Moreau (INSERM U618, Tours) for helpful discussion as we do for the following for financial support: the Ministère de la Recherche for supporting CM; INSERM; University François Rabelais; and Lions Club Jardin de France.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Michelle, C., Vourc’h, P., Mignon, L. et al. What Was the Set of Ubiquitin and Ubiquitin-Like Conjugating Enzymes in the Eukaryote Common Ancestor?. J Mol Evol 68, 616–628 (2009). https://doi.org/10.1007/s00239-009-9225-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00239-009-9225-6