Abstract
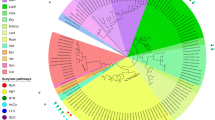
The human gut is extremely densely inhabited by bacteria mainly from two phyla, Bacteroidetes and Firmicutes, and there is a great interest in analyzing whole-genome sequences for these species because of their relation to human health and disease. Here, we do whole-genome comparison of 105 Bacteroidetes/Chlorobi genomes to elucidate their phylogenetic relationship and to gain insight into what is separating the gut living Bacteroides and Parabacteroides genera from other Bacteroidetes/Chlorobi species. A comprehensive analysis shows that Bacteroides species have a higher number of extracytoplasmic function σ factors (ECF σ factors) and two component systems for extracellular signal transduction compared to other Bacteroidetes/Chlorobi species. A whole-genome phylogenetic analysis shows a very little difference between the Parabacteroides and Bacteroides genera. Further analysis shows that Bacteroides and Parabacteroides species share a large common core of 1,085 protein families. Genome atlases illustrate that there are few and only small unique areas on the chromosomes of four Bacteroides/Parabacteroides genomes. Functional classification to clusters of othologus groups show that Bacteroides species are enriched in carbohydrate transport and metabolism proteins. Classification of proteins in KEGG metabolic pathways gives a detailed view of the genome’s metabolic capabilities that can be linked to its habitat. Bacteroides pectinophilus and Bacteroides capillosus do not cluster together with other Bacteroides species, based on analysis of 16S rRNA sequence, whole-genome protein families and functional content, 16S rRNA sequences of the two species suggest that they belong to the Firmicutes phylum. We have presented a more detailed and precise description of the phylogenetic relationships of members of the Bacteroidetes/Chlorobi phylum by whole genome comparison. Gut living Bacteroides have an enriched set of glycan, vitamin, and cofactor enzymes important for diet digestion.






Similar content being viewed by others
References
Bendtsen JD, Binnewies TT, Hallin PF, Sicheritz-Ponten T, Ussery DW (2005) Genome update: prediction of secreted proteins in 225 bacterial proteomes. Microbiology 151:1725–1727
Bendtsen JD, Binnewies TT, Hallin PF, Ussery DW (2005) Genome update: prediction of membrane proteins in prokaryotic genomes. Microbiology 151:2119–2121
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Binnewies TT, Bendtsen JD, Hallin PF, Nielsen N, Wassenaar TM, Pedersen MB, Klemm P, Ussery DW (2005) Genome update: protein secretion systems in 225 bacterial genomes. Microbiology 151:1013–1016
Carlier JP, Bedora-Faure M, K’Ouas G, Alauzet C, Mory F (2010) Proposal to unify Clostridium orbiscindens Winter et al. 1991 and Eubacterium plautii (Seguin 1928) Hofstad and Aasjord 1982, with description of Flavonifractor plautii gen. nov., comb. nov., and reassignment of Bacteroides capillosus to Pseudoflavonifractor capillosus gen. nov., comb. nov. Int J Syst Evol Microbiol 60:585–590
Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R (2009) Bacterial community variation in human body habitats across space and time. Science 326:1694–1697
Doolittle WF (1999) Phylogenetic classification and the universal tree. Science 284:2124–2129
Downes J, Sutcliffe IC, Hofstad T, Wade WG (2006) Prevotella bergensis sp. nov., isolated from human infections. Int J Syst Evol Microbiol 56:609–612
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Garrity GM, Lilburn TG, Cole JR, Harrison SH, Euzeby J, Tindall BJ (2007) Introduction to the Taxonomic Oultine of Bacteria and Archaea (TOBA) Release 7.7
Hallin PF, Binnewies TT, Ussery DW (2008) The genome BLASTatlas-a GeneWiz extension for visualization of whole-genome homology. Mol Biosyst 4:363–371
Hayashi H, Shibata K, Sakamoto M, Tomita S, Benno Y (2007) Prevotella copri sp. nov. and Prevotella stercorea sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 57:941–946
Hong SH, Kim TY, Lee SY (2004) Phylogenetic analysis based on genome-scale metabolic pathway reaction content. Appl Microbiol Biotechnol 65:203–210
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32:D277–280
Kiil K, Ferchaud JB, David C, Binnewies TT, Wu H, Sicheritz-Ponten T, Willenbrock H, Ussery DW (2005) Genome update: distribution of two-component transduction systems in 250 bacterial genomes. Microbiology 151:3447–3452
Kill K, Binnewies TT, Sicheritz-Ponten T, Willenbrock H, Hallin PF, Wassenaar TM, Ussery DW (2005) Genome update: sigma factors in 240 bacterial genomes. Microbiology 151:3147–3150
Kitahara M, Sakamoto M, Ike M, Sakata S, Benno Y (2005) Bacteroides plebeius sp. nov. and Bacteroides coprocola sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 55:2143–2147
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Lee ZM, Bussema C 3rd, Schmidt TM (2009) rrnDB: documenting the number of rRNA and tRNA genes in bacteria and archaea. Nucleic Acids Res 37:D489–493
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI (2005) Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 102:11070–11075
Ley RE, Peterson DA, Gordon JI (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848
Li L, Stoeckert CJ Jr, Roos DS (2003) OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI (2009) Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA 106:5859–5864
Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55:205–211
Nelson KE, Weinstock GM, Highlander SK, Worley KC, Creasy HH, Wortman JR, Rusch DB, Mitreva M, Sodergren E, Chinwalla AT, Feldgarden M, Gevers D, Haas BJ, Madupu R, Ward DV, Birren BW, Gibbs RA, Methe B, Petrosino JF, Strausberg RL, Sutton GG, White OR, Wilson RK, Durkin S, Giglio MG, Gujja S, Howarth C, Kodira CD, Kyrpides N, Mehta T, Muzny DM, Pearson M, Pepin K, Pati A, Qin X, Yandava C, Zeng Q, Zhang L, Berlin AM, Chen L, Hepburn TA, Johnson J, McCorrison J, Miller J, Minx P, Nusbaum C, Russ C, Sykes SM, Tomlinson CM, Young S, Warren WC, Badger J, Crabtree J, Markowitz VM, Orvis J, Cree A, Ferriera S, Fulton LL, Fulton RS, Gillis M, Hemphill LD, Joshi V, Kovar C, Torralba M, Wetterstrand KA, Abouellleil A, Wollam AM, Buhay CJ, Ding Y, Dugan S, FitzGerald MG, Holder M, Hostetler J, Clifton SW, Allen-Vercoe E, Earl AM, Farmer CN, Liolios K, Surette MG, Xu Q, Pohl C, Wilczek-Boney K, Zhu D (2010) A catalog of reference genomes from the human microbiome. Science 328:994–999
Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, Jin Z, Lee P, Yang L, Poles M, Brown SM, Sotero S, Desantis T, Brodie E, Nelson K, Pei Z (2010) Diversity of 16 S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol 76:3886–3897
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–120
Rautio M, Eerola E, Vaisanen-Tunkelrott ML, Molitoris D, Lawson P, Collins MD, Jousimies-Somer H (2003) Reclassification of Bacteroides putredinis (Weinberg et al., 1937) in a new genus Alistipes gen. nov., as Alistipes putredinis comb. nov., and description of Alistipes finegoldii sp. nov., from human sources. Syst Appl Microbiol 26:182–188
Sakamoto M, Benno Y (2006) Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol 56:1599–1605
Samuel BS, Gordon JI (2006) A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci USA 103:10011–10016
Shah HN, Collins DM (1990) NOTES: prevotella, a new genus to include bacteroides melaninogenicus and related species formerly classified in the genus bacteroides. Int J Syst Bacteriol 40:205–208
Shah HN, Collins MD (1988) Proposal for reclassification of bacteroides asaccharolyticus, bacteroides gingivalis, and bacteroides endodontalis in a New Genus, porphyromonas. Int J Syst Bacteriol 38:128–131
Shah HN, Collins MD (1989) Proposal to restrict the genus bacteroides (Castellani and Chalmers) to Bacteroides fragilis and closely related species. Int J Syst Bacteriol 39:85–87
Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69:183–215
Suzuki R, Shimodaira H (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tap J, Mondot S, Levenez F, Pelletier E, Caron C, Furet JP, Ugarte E, Munoz-Tamayo R, Paslier DL, Nalin R, Dore J, Leclerc M (2009) Towards the human intestinal microbiota phylogenetic core. Environ Microbiol 11:2574–2584
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinform 4:41
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI (2009) A core gut microbiome in obese and lean twins. Nature 457:480–484
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444:1027–1031
Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI (2003) A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076
Xu J, Chiang HC, Bjursell MK, Gordon JI (2004) Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol 12:21–28
Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, Martens EC, Henrissat B, Coutinho PM, Minx P, Latreille P, Cordum H, Van Brunt A, Kim K, Fulton RS, Fulton LA, Clifton SW, Wilson RK, Knight RD, Gordon JI (2007) Evolution of symbiotic bacteria in the distal human intestine. PLoS Biol 5:e156
Acknowledgements
We gratefully acknowledge the Knut and Alice Wallenberg Foundation and the Chalmers Foundation for financial support. We thank Dina Petranovic for helpful discussions for the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Bacteroidetes/Chlorobi genomes in this study. (RTF 332 kb)
Supplementary Figure 1
Transmembrane helices, sigma factors and two-component systems were predicted with hidden Markov models (HMM) with an e value cutoff of 0.01 [17, 18, 20], signal peptides were identified with SignalP 3.0 [3]. In each genome, the total number of features were counted. Mann–Whitney U test was used to calculate the probability of an equal distribution between Bacteroides, Parabacteroides, and other genomes, comparisons are indicated by B, P, and O, respectively. tRNAs total number of tRNA genes, ecf ECF-sigma factors, HisKA_1 two-component histidine kinase 1, RRreciever two-component response regulator, Total total number of proteins, CYT cytosolic proteins, SpI proteins with signal peptidase I cleavage site, SpII proteins with signal peptidase II cleavage site, TMH proteins with at least one trans-membrane helix. (PDF 30 kb)
Rights and permissions
About this article
Cite this article
Karlsson, F.H., Ussery, D.W., Nielsen, J. et al. A Closer Look at Bacteroides: Phylogenetic Relationship and Genomic Implications of a Life in the Human Gut. Microb Ecol 61, 473–485 (2011). https://doi.org/10.1007/s00248-010-9796-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00248-010-9796-1