Abstract
The HLA-A locus represents a single copy gene that displays abundant allelic polymorphism in the human population, whereas, in contrast, a nonhuman primate species such as the rhesus macaque (Macaca mulatta) possesses multiple HLA-A-like (Mamu-A) genes, which parade varying degrees of polymorphism. The number and combination of transcribed Mamu-A genes present per chromosome display diversity in a population of Indian animals. At present, it is not clearly understood whether these different A region configurations are evolutionarily stable entities. To shed light on this issue, rhesus macaques from a Chinese population and a panel of cynomolgus monkeys (Macaca fascicularis) were screened for various A region-linked variations. Comparisons demonstrated that most A region configurations are old entities predating macaque speciation, whereas most allelic variation (>95%) is of more recent origin. The latter situation contrasts the observations of the major histocompatibility complex class II genes in rhesus and cynomolgus macaques, which share a high number of identical alleles (>30%) as defined by exon 2 sequencing.
Similar content being viewed by others
Introduction
Major histocompatibility complex (MHC) represents a multigene family that plays a crucial role in the generation of adaptive immune responses in vertebrate species. A key feature of the system is that some of its genes display abundant polymorphism at the population level. In addition, the number of Mhc class I or II genes may differ significantly between species, as well as between individuals of a species (Kelley et al. 2005). MHC polymorphisms may have a profound impact on several features such as disease susceptibility, organ transplantation, and reproductive success (Lagaaij et al. 1989; Goulder and Watkins 2004; Bontrop and Watkins 2005; Ziegler et al. 2005; Smith et al. 2006). The MHC systems of various primate species, including humans, have been studied extensively (Watkins 1995; Antunes et al. 1998; Bontrop et al. 1999; Adams and Parham 2001; de Groot et al. 2002; Lafont et al. 2004; Middleton et al. 2004; Marsh et al. 2005; Penedo et al. 2005; Abbott et al. 2006; Huchard et al. 2006). For example, the MHC of the rhesus macaque (MhcMamu), an Old World primate species, has been shown to share many similarities with the human leukocyte antigen (HLA) system in humans. The evolutionary orthologs of the HLA-DP, -DQ, and -DR genes are also present in rhesus monkeys, and as in humans, these loci are polymorphic (Bontrop et al. 1999). The number of Mamu-DRB region configurations appears to be expanded in comparison with humans, and some of these regions seem to harbor an extended number of genes (Doxiadis et al. 2000). Subsequent cDNA studies have illustrated, however, that in humans and rhesus macaques, comparable numbers of DRB genes are transcribed (de Groot et al. 2004).
Apart from Mamu-E (Knapp et al. 1998), -F (Otting and Bontrop 1993), -G (Boyson et al. 1996a; Castro et al. 1996), and -AG (Boyson et al. 1997), rhesus macaques also possess a B-like sequence, designated Mamu-I, which appears to be present in each haplotype (Urvater et al. 2000). As observed in humans, these nonclassical genes display low levels of polymorphism. An ortholog of the HLA-C gene appears to be absent in the rhesus macaque, whereas evolutionary equivalents of the classical HLA-A and -B genes have been described (Boyson et al. 1996b). The Mamu-A and -B genes have been subjected to several rounds of duplication as was shown by genomic sequencing (Daza-Vamenta et al. 2004; Kulski et al. 2004). Analysis of a panel of rhesus macaques, mainly originating from the Indian subcontinent, illustrated that the number and combination of Mamu-A and -B genes that are expressed per haplotype may differ at the population level (Otting et al. 2005). In the same study, marked differences in expression levels were shown. At this stage it is not known whether rhesus macaques originating from other geographic areas have unique Mhc class I alleles and/or region configurations. Comparative studies have illustrated that many Mhc loci and lineages predate speciation events. The sharing of Mhc alleles between two primate species seems to be a rare event, and only a few cases have been documented (Cooper et al. 1998; Evans et al. 1998). An exception is provided by rhesus and cynomolgus macaques, which seem to share a high number of Mhc class II alleles as was defined by exon 2 sequencing (Blancher et al. 2006; Doxiadis et al. 2006). Whether this sharing is because of introgression or purifying selection remains to be elucidated. Thus far, about 50 Mafa-A sequences have been published for the cynomolgus macaque (Uda et al. 2004; Krebs et al. 2005). The absence of pedigreed material did not allow us to define Mafa-A loci or region configurations. For that reason, a panel of pedigreed cynomolgus macaques was incorporated in the present study. Comparison with rhesus macaque Mhc class I sequences obtained from different populations enabled us to draw conclusions on the evolutionary stability of A region variations.
Materials and methods
Animals and cell lines
The Biomedical Primate Research Centre houses a self-sustaining colony of approximately 1,000 rhesus macaques, mainly of Indian origin, that have been pedigreed based on the segregation of serologically defined MHC allotypes (Bontrop et al. 1999; Penedo et al. 2005; Doxiadis et al. 2006). Furthermore, a large collection of DNA samples as well as B-cell lines is available. In this study, B-cells derived from rhesus macaques of Chinese origin and from cynomolgus monkeys were used to isolate RNA. Most cynomolgus monkey samples originate from a pedigreed group housed at the campus of the University of Utrecht.
cDNA cloning and sequencing
RNA was isolated from B cells (Rneasy kit, Qiagen) and subjected to One-step reverse transcriptase polymerase chain reaction (RT-PCR), as recommended by the supplier (Qiagen). The primers (5′MAS) AATTCATGGCGCCCCGAACCCTCCTCCTGG, and (3′MAS) CTAGACCACACAAGGCGGCTGTCTCAC were used, which are specific for class I A transcripts in macaques. The final elongation step was extended to 30 min to generate a 3′dA overhang. The RT-PCR products were cloned using the InsT/Aclone kit (Fermentas). After transformation, 16 to 32 colonies were picked for plasmid isolations. The separate Mamu-A loci show differences in expression levels, as indicated by the numbers of clones, picked from each animal. The alleles with high expression levels (majors) may be associated with serotypes and thus involved in classical antigen presentation, whereas those with low expression levels (minors) are considered nonclassicals and may exhibit more specialized types of functions (Otting et al. 2005).
Sequencing reactions were performed using the BigDye terminator cycle sequencing kit, and samples were run on an automated capillary sequencing system (ABI Genetic Analyzer 3100) as has been described previously (Otting et al. 2005).
Locus-specific PCR reactions
To test the presence of Mamu- and Mafa-A4*14 alleles, 1 μl of the RT-PCR samples was taken for a single-specific-primer PCR (SSP-PCR) reaction in advance of the cloning step. The relevant primers, (5′A*14) GGGACCCGACGGGCGCCTCCAA and (3′A*14) GGCCCTCCAGGTAGACTCTGTC have annealing sites in exon 3. Amplifications were carried out starting with 2 min at 94°C, followed by 25 cycles at 94°C, 65°C, and 72°C for 1 min each. The PCR products were subjected to direct sequencing, and the reactions were performed as described above.
Phylogenetic analysis and nomenclature
The sequences were analyzed with the Sequence Navigator Software version 1.0.1 (Applied Biosystems), and alleles are based on at least three clones with identical full-length sequences. To define loci and lineages, alignments of the sequences were made using the MacVector™ version 8.1.1 (Oxford Molecular Group), followed by manual adjustments. Phylogenetic analyses on the full-length (1,065 bp) sequences were also performed with the MacVector software. Neighbor-joining trees were constructed with the Kimura 2 parameter method. Bootstrap analyses were performed based on 1,000 replications.
In total, 130 unreported Mamu-A and Mafa-A sequences were submitted to the European Bioinformatics Institute and European Molecular Biology Laboratory (EBI-EMBL) database. Relevant information such as accession number and a reference cell-line are provided (Table 1). Moreover, all novel Mamu-A and Mafa-A sequences are named in accordance with a generally accepted nomenclature proposal (Klein et al. 1990; Robinson et al. 2003; Ellis et al. 2006). For example, Mamu-A1*0101 defines a Mhc allele in the rhesus macaque, which is encoded by one of the class I loci: namely, A1. The first two digits after the asterisk define the lineage, whereas the third and fourth digits define the allele number. These allele numbers are arbitrary, as they reflect the order in which the alleles were discovered. A fifth and sixth digit are used to mark a synonymous basepair difference between two sequences.
Results and discussion
Polymorphism and diversity of the Mamu-A region: comparison of Chinese and Indian rhesus macaques
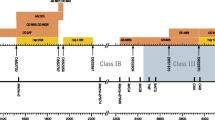
In a previous communication, five different Mamu-A region configurations were defined in a population of Indian rhesus macaques (Otting et al. 2005). These configurations display diversity with regard to the number and combination of distinct Mamu-A genes present per chromosome. The loci have been designated Mamu-A1, -A2, -A3, and -A4, respectively (Fig. 1). As can be seen, each region configuration comprises a Mamu-A1 gene characterized by high transcription levels (major) combined with one or two other Mamu-A genes characterized by lower transcription levels (minors). The Mamu-A1 gene is probably responsible for executing the classical antigen presentation function (Evans et al. 1999; Sidney et al. 2000; Sette et al. 2005), whereas the others can be considered as nonclassicals and may interact, for instance, with the KIR gene family present on NK cells. Two complete MHC haplotypes in the rhesus macaque are sequenced (Daza-Vamenta et al. 2004; Kulski et al. 2004), and they represent two-region configurations. One haplotype contains the -A1/-A2 combination, whereas in the other, the -A1/-A4 pair is observed. Both region configurations are present in the animals of Indian origin (Fig. 1).
Schematic representation of different Mamu-A region configurations observed in Indian rhesus macaques. The exact order and physical distances of the loci on the genome are still unknown. The relative levels of polymorphism and transcription of the loci are indicated. Two Mamu-A region configurations (1 and 5) are confirmed by the sequencing of the complete rhesus MHC region (Daza-Vamenta et al. 2004; Kulski et al. 2004)
Within the present panel of 42 Chinese animals, 59 different full-length Mamu-A cDNAs were detected that can be grouped into various loci and lineages (Fig. 2). A complete listing of the alleles detected in the Chinese macaques is provided as electronic supplementary material (Table 5). Because of codominant expression, rhesus macaques can be heterozygous for the Mamu-A1 gene and thus can express up to two allotypes. Nine animals were found to express three different Mamu-A1-like sequences. These animals shared one of these sequences in separated clades in phylogenetic tree (Fig. 2; -A5, -A6, and -A7). The triplets of the three animals are indicated by asterisks. The clustering of the three alleles, obtained from one animal, in clades distinct from -A2, -A3, and -A4 indicates that one allele belongs to a separate locus. The newly detected loci are named Mamu-A5, -A6, and -A7, according to the generally accepted nomenclature system (Klein et al. 1990; Robinson et al. 2003; Ellis et al. 2006). Representatives of two loci, A5 and A6, are also observed in cynomolgus macaques that express more than two A1-like sequences.
Phylogenetic tree of Mamu-A and Mafa-A gene/alleles detected in this study. The -A2 locus is represented by four alleles only. The tree is based on full-length sequences, although analyses on only exon 2 and 3 showed no significant differences. The nomenclature of the new alleles is based on a more extended tree (not shown) containing all the Mamu-A and Mafa-A alleles now available. Asterisk, three alleles of animal Ri081, of which one represents the locus Mamu-A5. Double asterisks, three alleles of animal Ri145, of which one represents the locus Mamu-A6. Triple asterisks, three alleles of animal Ri078, of which one represents the locus Mamu-A7
Thirty-three new Mamu-A1 alleles were detected in the Chinese rhesus macaques. Although some of the alleles in this population group into lineages that are also present in Indian animals, sharing of alleles for the Mamu-A1 locus is rare and was observed only once. The Mamu-A1*26 allele is detected in both populations, whereas the Mamu-A1*03 and for -A1*04, alleles are highly similar and differ for one synonymous basepair substitution. Hence, it is concluded that most allelic polymorphism observed for the Mamu-A1 locus was probably generated after the rhesus macaque populations were separated. Sequence comparisons illustrated that most of the variations map at the contact residues of the peptide-binding site (data not shown). Thus, polymorphism at the highly divergent Mamu-A1 locus, characterized by high expression levels, must have resulted from positive Darwinian selection (Hughes and Nei 1988; Borghans et al. 2004).
The Mamu-A2 gene displays differential haplotype distribution in the Indian population of rhesus macaques (Fig. 1). This also appears to be the case for Chinese animals, as Mamu-A2 cDNAs were detected in 39 out of 42 of them. In this panel, 16 alleles that differ from the ones earlier detected in Indian animals were defined. In the Indian population, low expression levels characterize the Mamu-A2 gene, which appears also to be the case in Chinese animals.
Five Chinese macaques possess the Mamu-A3*1307 sequence, which is always observed in combination with Mamu-A1*1102. In concordance with the Indian animals, the number of Mamu-A3 clones detected also reflects low expression levels. A full-length Mamu-A4*1404 cDNA was detected in only three animals. In contrast to the Mamu-A4 allele in the Indian macaque, the Chinese allele has a stopcodon at the last triplet of exon 5 encoding the transmembrane part of the class I protein. This suggests that the corresponding gene product may settle in the membrane, but the signal transduction is impaired. As was observed earlier in Indian animals, this locus is characterized by extremely low expression levels (Fig. 1). For that reason, the presence of Mamu-A4 cDNAs in the other animals was tested with SSP on the RT-PCR samples, and 17 out of 42 Chinese animals appeared to be positive for this locus. The Mamu-A5, -A6, and -A7 genes are also characterized by low expression levels and are considered to represent minors.
In contrast to the previously studied Indian population, the Chinese animals were selected randomly from a large population. Because of the lack of pedigree data and segregation profiles, it was not possible to firmly establish the segregation of different Mamu-A genes present per chromosome. Haplotypes can only be deduced based on sharing of sequences between animals, and such haplotypes are listed in Table 2. At this stage, it is impossible to determine in which region configurations the Mamu-A5, -A6, and -A7 genes are present.
Mafa-A region polymorphism and diversity in cynomolgus monkeys
Sixty-two alleles were detected in the 96 cynomolgus macaques analyzed. As observed in the rhesus macaques, most animals had two polymorphic Mafa-A1 sequences, in combination with orthologs of the Mamu-A2, -A3, -A4, -A5, and/or -A6 genes. In total, 38 Mafa-A1 alleles were detected, illustrating the high level of polymorphism of this locus. These alleles were compared with sequences reported earlier by two other research groups (Uda et al. 2004; Krebs et al. 2005) that have used different nomenclature systems. Only four alleles -A1*3101, -A1*3201, -A1*3801, and -A5*3001 detected in our panel are identical to the earlier described sequences A*310101, A*320101, A*380101, and A*300101.
A total of 79 out of 96 animals have at least one Mafa-A2 sequence, and 18 alleles could be distinguished (Table 1). The oligomorphic A2 locus is also present in the pigtailed macaque (Macaca nemestrina) as was recently described (Pratt et al. 2006; Lafont et al. 2007). In these studies, 90% of the animals possess the gene, and lower transcription levels were also observed in comparison to the other Mane-A alleles. The Mafa-A3 gene is observed in only two animals, whereas the Mafa-A4 locus is detected in 41 individuals. Two Mafa-A4 alleles were found that differ in only one basepair (nonsynonymous) substitution (Fig. 2). As in the Chinese rhesus macaques, the Mafa-A4 alleles have a stopcodon at the end of exon 5. The Mafa-A5 and -A6 alleles were detected in, respectively, eight and four animals and display low levels of polymorphism. Phylogenetic analyses illustrate that apart from loci rhesus and cynomolgus monkeys also share lineages (Fig. 2). As found in rhesus macaques, the Mafa-A1 alleles are characterized by high transcription levels, whereas the other loci display moderate or low expression levels.
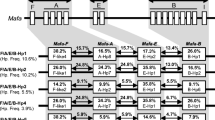
Because most of the cynomolgus macaques are pedigreed, the combination of sequences that are inherited on one chromosome could be defined. As an example, the kinship tree of one breeding group is provided (Fig. 3). The haplotypes are grouped based on the combination of loci (Table 3). Seven combinations of loci or region configurations are recognized (Fig. 4), and four of them (two to five) are also observed in the Indian rhesus macaques (Fig. 1), which indicates that they predate speciation. Mafa-A5*3001 segregates in combination with Mafa-A1*6001 and -A1*7201 but not exclusively. The exact region configurations of Mafa-A6 containing haplotypes are not yet known.
Schematic representation of different Mafa-A region configurations in cynomolgus macaques. Four configurations (2–5) are shared with the Indian rhesus macaques (Fig. 1). One region configuration lacks the Mafa-A1 gene. It is not yet known whether the locus is really absent or missed because of PCR failure. Should a Mafa-A1 gene be found in extended studies than this region, configuration has to be deleted from the list
Sharing of -A1 and -A2 sequences between different macaque species
A comparison of all available Mamu-A to Mafa-A sequences showed that six full-length cDNA transcripts are shared between both species of macaque. This is the case for five -A1 pairs and one -A2 pair (Table 4). One of these cynomolgus monkey alleles is identical to an Indian rhesus allele, whereas the other five are shared with animals of Chinese origin. This observation was unexpected as the two rhesus macaque populations investigated share only one allele. The fact that cynomolgus macaques share more alleles with Chinese rhesus macaques than with Indian individuals may be explained by the fact that there is an overlap in the geographic areas inhabited by both species in Indochina (eastern Asia). It is known that rhesus macaques and cynomolgus monkeys can interbreed and produce offspring (Tosi et al. 2002). Sharing of alleles was not observed for the minors controlled by the A3–A7 loci.
Different modes of selection operating on the Mhc class I and II genes in macaques
Although Mamu-A and Mafa-A sequences are interspersed in the phylogenetic tree, the vast majority of the alleles are species unique, and for rhesus macaque, most of them appear to be population specific. This is in sharp contrast to the situation observed for the macaque class II region, where sharing for different exon 2 sequences at the Mhc-DR, but especially the -DQ and -DP genes, is far more common (Blancher et al. 2006; Doxiadis et al. 2006). About half of the Mafa-DPB1, -DQA1, and -DQB1 and one third of the Mafa-DRB sequences are identical to rhesus orthologs. As animals of the same populations have been used to study Mhc class I and II sequences, the present results exclude the possibility that this high sharing of Mhc class II sequences is because of introgression. A more likely explanation is that in macaques, the exon 2 sequences of the Mhc class II genes, which encode the peptide-binding site, have been subjected to purifying selection. As a consequence, many Mhc class II alleles in these two macaque species predate speciation processes. A databank search indicated that the phenomenon also extends to other macaque species (Robinson et al. 2003).
The Mhc class I alleles in macaques are largely unique, illustrating that selection has favored diversity. Mhc class I proteins are involved in the presentation of intracellular pathogens such as viruses and parasites that are known to evolve at high mutation rates. Recognition of Mhc class I molecules combined with a foreign peptide may result in the lysis of an infected cell. The Mhc class II molecules select peptides, originating from extracellular pathogens such as bacteria and fungi, for binding. Mhc class II-mediated activation may result in antibody production and/or providing help to cytotoxic T cells. The present results illustrate that, probably because of coevolution with intracellular pathogens, macaques have generated a highly complex and divergent Mhc class I repertoire. The fact that Mhc class I alleles evolve quickly, even within a species, has been documented in humans for instance (Belich et al. 1992; Watkins et al. 1992). The high degree of sharing of Mhc class II sequences seems to be unique for macaques and has not been observed in any other group of vertebrate species.
References
Abbott KM, Wickings EJ, Knapp LA (2006) High levels of diversity characterize mandrill (Mandrillus sphinx) Mhc-DRB sequences. Immunogenetics 58:628–640
Adams EJ, Parham P (2001) Species-specific evolution of MHC class I genes in the higher primates. Immunol Rev 183:41–64
Antunes SG, de Groot NG, Brok H, Doxiadis G, Menezes AA, Otting N, Bontrop RE (1998) The common marmoset: a new world primate species with limited Mhc class II variability. Proc Natl Acad Sci USA 95:11745–11750
Belich MP, Madrigal JA, Hildebrand WH, Zemmour J, Williams RC, Luz R, Petzl-Erler ML, Parham P (1992) Unusual HLA-B alleles in two tribes of Brazilian Indians. Nature 357:326–329
Blancher A, Tisseyre P, Dutaur M, Apoil PA, Maurer C, Quesniaux V, Raulf F, Bigaud M, Abbal M (2006) Study of Cynomolgus monkey (Macaca fascicularis) MhcDRB (Mafa-DRB) polymorphism in two populations. Immunogenetics 58:269–282
Bontrop RE, Watkins DI (2005) MHC polymorphism: AIDS susceptibility in non-human primates. Trends Immunol 26:227–233
Bontrop RE, Otting N, de Groot NG, Doxiadis GG (1999) Major histocompatibility complex class II polymorphisms in primates. Immunol Rev 167:339–350
Borghans JA, Beltman JB, De Boer RJ (2004) MHC polymorphism under host–pathogen coevolution. Immunogenetics 55:732–739
Boyson JE, Iwanaga KK, Golos TG, Watkins DI (1996a) Identification of the rhesus monkey HLA-G ortholog. Mamu-G is a pseudogene. J Immunol 157:5428–5437
Boyson JE, Shufflebotham C, Cadavid LF, Urvater JA, Knapp LA, Hughes AL, Watkins DI (1996b) The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J Immunol 156:4656–4665
Boyson JE, Iwanaga KK, Golos TG, Watkins DI (1997) Identification of a novel MHC class I gene, Mamu-AG, expressed in the placenta of a primate with an inactivated G locus. J Immunol 159:3311–3321
Castro MJ, Morales P, Fernandez-Soria V, Suarez B, Recio MJ, Alvarez M, Martin-Villa M, Arnaiz-Villena A (1996) Allelic diversity at the primate Mhc-G locus: exon 3 bears stop codons in all Cercopithecinae sequences. Immunogenetics 43:327–336
Cooper S, Adams EJ, Wells RS, Walker CM, Parham P (1998) A major histocompatibility complex class I allele shared by two species of chimpanzee. Immunogenetics 47:212–217
Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE (2004) Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res 14:1501–1515
de Groot NG, Otting N, Doxiadis GG, Balla-Jhagjhoorsingh SS, Heeney JL, van Rood JJ, Gagneux P, Bontrop RE (2002) Evidence for an ancient selective sweep in the MHC class I gene repertoire of chimpanzees. Proc Natl Acad Sci USA 99:11748–11753
de Groot N, Doxiadis GG, De Groot NG, Otting N, Heijmans C, Rouweler AJ, Bontrop RE (2004) Genetic makeup of the DR region in rhesus macaques: gene content, transcripts, and pseudogenes. J Immunol 172:6152–6157
Doxiadis GG, Otting N, de Groot NG, Noort R, Bontrop RE (2000) Unprecedented polymorphism of Mhc-DRB region configurations in rhesus macaques. J Immunol 164:3193–3199
Doxiadis GG, Rouweler AJ, de Groot NG, Louwerse A, Otting N, Verschoor EJ, Bontrop RE (2006) Extensive sharing of MHC class II alleles between rhesus and cynomolgus macaques. Immunogenetics pp 1–10
Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, Stet RJ, Waller MJ, Walter L, Marsh SG (2006) ISAG/IUIS-VIC comparative MHC nomenclature committee report, 2005. Immunogenetics 57:953–958
Evans DT, Piekarczyk MS, Cadavid L, Hinshaw VS, Watkins DI (1998) Two different primate species express an identical functional MHC class I allele. Immunogenetics 47:206–211
Evans DT, O’Connor DH, Jing P, Dzuris JL, Sidney J, da Silva J, Allen TM, Horton H, Venham JE, Rudersdorf RA, Vogel T, Pauza CD, Bontrop RE, DeMars R, Sette A, Hughes AL, Watkins DI (1999) Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat Med 5:1270–1276
Goulder PJ, Watkins DI (2004) HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol 4:630–640
Huchard E, Cowlishaw G, Raymond M, Weill M, Knapp LA (2006) Molecular study of Mhc-DRB in wild chacma baboons reveals high variability and evidence for trans-species inheritance. Immunogenetics 58:805–816
Hughes AL, Nei M (1988) Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature 335:167–170
Kelley J, Walter L, Trowsdale J (2005) Comparative genomics of major histocompatibility complexes. Immunogenetics 56:683–695
Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI (1990) Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics 31:217–219
Knapp LA, Cadavid LF, Watkins DI (1998) The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J Immunol 160:189–196
Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O’Connor DH (2005) Unusually high frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239
Kulski JK, Anzai T, Shiina T, Inoko H (2004) Rhesus macaque class I duplicon structures, organization, and evolution within the alpha block of the major histocompatibility complex. Mol Biol Evol 21:2079–2091
Lafont BA, Buckler-White A, Plishka R, Buckler C, Martin MA (2004) Pig-tailed macaques (Macaca nemestrina) possess six MHC-E families that are conserved among macaque species: implication for their binding to natural killer receptor variants. Immunogenetics 56:142–154
Lafont BAP, McGraw CM, Stukes SA, Buckler-White A, Plishka RJ, Hirsch VM, Martin MA (2007) The locus encoding an oligomorphic family of MHC-A alleles (Mane-A*06/Mamu-A*05) is present at high frequency in several macaque species. Immunogenetics 59:211–223
Lagaaij EL, Hennemann IP, Ruigrok M, de Haan MW, Persijn GG, Termijtelen A, Hendricks GF, Weimar W, Claas FH, van Rood JJ (1989) Effect of one-HLA-DR-antigen-matched and completely HLA-DR-mismatched blood transfusions on survival of heart and kidney allografts. N Engl J Med 321:701–705
Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J (2005) Nomenclature for factors of the HLA System, 2004. Hum Immunol 66:571–636
Middleton SA, Anzenberger G, Knapp LA (2004) Identification of New World monkey MHC-DRB alleles using PCR, DGGE and direct sequencing. Immunogenetics 55:785–790
Otting N, Bontrop RE (1993) Characterization of the rhesus macaque (Macaca mulatta) equivalent of HLA-F. Immunogenetics 38:141–145
Otting N, Heijmans CM, Noort RC, de Groot NG, Doxiadis GG, van Rood JJ, Watkins DI, Bontrop RE (2005) Unparalleled complexity of the MHC class I region in rhesus macaques. Proc Natl Acad Sci USA 102:1626–1631
Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, de Groot N, de Groot NG, Ward T, Doxiadis GG (2005) Microsatellite typing of the rhesus macaque MHC region. Immunogenetics 57:198–209
Pratt BF, O’Connor DH, Lafont BA, Mankowski JL, Fernandez CS, Triastuti R, Brooks AG, Kent SJ, Smith MZ (2006) MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics 58:995–1001
Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG (2003) IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res 31:311–314
Sette A, Sidney J, Bui HH, del Guercio MF, Alexander J, Loffredo J, Watkins DI, Mothe BR (2005) Characterization of the peptide-binding specificity of Mamu-A*11 results in the identification of SIV-derived epitopes and interspecies cross-reactivity. Immunogenetics 57:53–68
Sidney J, Dzuris JL, Newman MJ, Johnson RP, Kaur A, Amitinder K, Walker CM, Appella E, Mothe B, Watkins DI, Sette A (2000) Definition of the Mamu-A*01 peptide binding specificity: application to the identification of wild-type and optimized ligands from simian immunodeficiency virus regulatory proteins. J Immunol 165:6387–6399
Smith WP, Vu Q, Li SS, Hansen JA, Zhao LP, Geraghty DE (2006) Toward understanding MHC disease associations: Partial resequencing of 46 distinct HLA haplotypes. Genomics
Tosi AJ, Morales JC, Melnick DJ (2002) Y-chromosome and mitochondrial markers in Macaca fascicularis indicate introgression with Indochinese M. mulatta and a biogeographic barrier in the Isthmus of Kra. Int J Primatol 23:161–178
Uda A, Tanabayashi K, Yamada YK, Akari H, Lee YJ, Mukai R, Terao K, Yamada A (2004) Detection of 14 alleles derived from the MHC class I A locus in cynomolgus monkeys. Immunogenetics 56:155–163
Urvater JA, Otting N, Loehrke JH, Rudersdorf R, Slukvin, II, Piekarczyk MS, Golos TG, Hughes AL, Bontrop RE, Watkins DI (2000) Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J Immunol 164:1386–1398
Watkins DI (1995) The evolution of major histocompatibility class I genes in primates. Crit Rev Immunol 15:1–29
Watkins DI, McAdam SN, Liu X, Strang CR, Milford EL, Levine CG, Garber TL, Dogon AL, Lord CI, Ghim SH et al (1992) New recombinant HLA-B alleles in a tribe of South American Amerindians indicate rapid evolution of MHC class I loci. Nature 357:329–333
Ziegler A, Kentenich H, Uchanska-Ziegler B (2005) Female choice and the MHC. Trends Immunol 26:496–502
Acknowledgment
The authors wish to thank Ms. Donna Devine for editing the manuscript and Henk van Westbroek for preparing the figures.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study was supported in part by the National Institutes of Health, project 1-R24-RR16038-01 (Catalog of Federal Assistance 93.306) and by the NIH/NCRR project U24-RR18144-01.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table 5
Overview of Mamu-A genes/alleles in the panel of Chinese rhesus macaques. Sequences represented by two ciphers (for example A*05) are not confirmed and are not submitted to the to the EBI-EMBL database (PDF 127 KB).
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Otting, N., de Vos-Rouweler, A.J.M., Heijmans, C.M.C. et al. MHC class I A region diversity and polymorphism in macaque species. Immunogenetics 59, 367–375 (2007). https://doi.org/10.1007/s00251-007-0201-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-007-0201-2