Abstract
The human major histocompatibility complex (MHC) is contained within about 4 Mb on the short arm of chromosome 6 and is recognised as the most variable region in the human genome. The primary aim of the MHC Haplotype Project was to provide a comprehensively annotated reference sequence of a single, human leukocyte antigen-homozygous MHC haplotype and to use it as a basis against which variations could be assessed from seven other similarly homozygous cell lines, representative of the most common MHC haplotypes in the European population. Comparison of the haplotype sequences, including four haplotypes not previously analysed, resulted in the identification of >44,000 variations, both substitutions and indels (insertions and deletions), which have been submitted to the dbSNP database. The gene annotation uncovered haplotype-specific differences and confirmed the presence of more than 300 loci, including over 160 protein-coding genes. Combined analysis of the variation and annotation datasets revealed 122 gene loci with coding substitutions of which 97 were non-synonymous. The haplotype (A3-B7-DR15; PGF cell line) designated as the new MHC reference sequence, has been incorporated into the human genome assembly (NCBI35 and subsequent builds), and constitutes the largest single-haplotype sequence of the human genome to date. The extensive variation and annotation data derived from the analysis of seven further haplotypes have been made publicly available and provide a framework and resource for future association studies of all MHC-associated diseases and transplant medicine.
Similar content being viewed by others
Introduction
The MHC has long been believed to be the most important region in the human genome with respect to infection, inflammation, autoimmunity and transplant medicine (Lechler and Warrens 2000). This was recently confirmed by the largest genome-wide association study carried out to date for seven common diseases, including two autoimmune diseases (type 1 diabetes and rheumatoid arthritis) and one inflammatory disease (Crohn’s disease). The highest associations were found between the MHC and these two autoimmune diseases (The Wellcome Trust Case Control Consortium 2007). The complex aetiology of MHC-associated disease coupled with high density, polymorphism, linkage disequilibrium (LD) and frequent non-Mendelian inheritance of gene loci have made it challenging to identify variations that cause or contribute to disease phenotypes. Additional limiting factors have been our incomplete knowledge of the allelic variation of genes and regions flanking the nine classical human leukocyte antigen (HLA) loci and the lack of a single haplotype reference sequence, the original reference sequence being a composite of multiple MHC haplotypes (Mungall et al. 2003; The MHC Sequencing Consortium 1999).
Recognizing that the future identification of variants conferring susceptibility to common disease is critically dependent on fully informative polymorphism and haplotype maps, the MHC Haplotype Consortium formed in 2000 with the aim to generate these critical data and to make them publicly available as a general resource for MHC-linked disease studies. Similar efforts, but with different experimental approaches, were also carried out in Japan (Shiina et al. 2006) and the USA (Smith et al. 2006). To develop the resource, eight HLA-homozygous MHC haplotypes were selected on the basis of conferring either protection against or susceptibility to two autoimmune diseases, type 1 diabetes and multiple sclerosis, and that represented common haplotypes in European populations. In the subsequent years, incremental data, materials and tools comprising this resource have been released (Allcock et al. 2002; Horton et al. 2004; Stewart et al. 2004; Traherne et al. 2006) and have contributed towards the construction of a high-resolution LD map and a first generation of HLA tag single nucleotide polymorphisms (SNPs; de Bakker et al. 2006; Miretti et al. 2005) and the identification of a second MHC susceptibility locus for multiple sclerosis (The International Multiple Sclerosis Genetics Consortium; Yeo et al. 2007). In this paper, we report the final account of this international effort, including, analysis of the last four of the eight haplotypes, up-to-date variation statistics, gene annotation, population-specific aspects and a detailed description of the databases and tools for viewing and accessing the data in the context of existing genome annotation.
Materials and methods
Variation analysis
The method previously reported for comparison of MHC haplotype sequences (Stewart et al. 2004; Traherne et al. 2006) was extended to cover all eight haplotypes. Briefly, the most suitable method proved to be a clone by clone comparison using the discrepancy-list option of the cross_match program (Green, unpublished; http://www.phrap.org/), an implementation of the Smith–Waterman sequence alignment algorithm (Smith and Waterman 1981), using the alignment of a haplotype clone sequence with the appropriate overlapping reference sequence from a PGF clone or clones. All variations were submitted to dbSNP using the submitter handle SI_MHC_SNP and user identifiers of the form [PGF BAC clone sequence version]_[position in PGF BAC clone sequence]_[variation change]. Thus, AL662890.3_6645_TC indicates a substitution in which the base T at base position 6645 in AL662890.3 (PGF BAC 308K3) was substituted by C in the other haplotype. In the case of indels, the ‘variation change’ consists of ‘i’ or ‘d’ (for insertion or deletion), followed by a numerical value for the length of the indel, in turn followed by the inserted or deleted sequence if this were of 12 or fewer bases. For longer indels, an X value is given, which refers to a look-up table (http://www.sanger.ac.uk/HGP/Chr6/MHC/Xfile). Thus, AL662890.3_7470_d8TACACACA indicates a deletion in AL662890.3 after base 7470 of the eight bases ‘TACACACA’. Further, AL662890.3_10559_i5ATATT indicates an insertion in AL662890.3 starting after base 10559 of the five bases ‘ATATT’. AL662890.3_7475_d14X1 indicates a 14-base deletion after base 7475 in AL662890.3 of a sequence coded as X1 which is ‘ATACACACACACAC’.
Major indel sequences, appearing as breaks in the cross_match discrepancy lists between two clones from difference haplotypes, were extracted and subjected to analysis by RepeatMasker to detect the presence of retrotransposible elements.
Gene annotation
The finished genomic sequence for each of the eight haplotypes was analysed using a modified Ensembl pipeline (Searle et al. 2004). CpG islands were predicted on unmasked sequence. Interspersed and tandem repeats were masked out by RepeatMasker (Smit, AFA, Hubley, R & Green, P. RepeatMasker Open-3.0. 1996–2004, http://www.repeatmasker.org) and Tandem Repeats Finder (TRF; Benson 1999), respectively. The sequence was then BLAST searched (BLAST, basic local alignment search tool; Altschul et al. 1990) using a vertebrate set of complementary DNAs (cDNAs) and expressed sequence tags (ESTs) from the European Molecular Biology Laboratory (EMBL) nucleotide database (Kulikova et al. 2007), followed by the re-alignment of significant hits. Non-redundant proteins were aligned similarly. Protein domain matches were provided through alignment of Pfam to the genomic sequence using Genewise (Birney et al. 2004), thereby providing protein domain data to the annotator. Ab initio gene predictions were performed by Genscan (Burge and Karlin 1997) and Fgenesh (Salamov and Solovyev 2000), and potential transcriptional start sites were predicted by Eponine (Down and Hubbard 2002). Analysis results were displayed, and annotation was performed through an in-house annotation software system.
Genes were manually annotated according to the human and vertebrate analysis and annotation (HAVANA) guidelines (http://www.sanger.ac.uk/HGP/havana/) using evidence based on comparison with external databases as of August 2005. All gene structures are supported by transcriptional evidence, either from cDNA, EST, or protein. In general, annotations are supported by best-in-genome evidence. Haplotype-specific evidence is assigned where possible. As with previous MHC annotation (Stewart et al. 2004; Traherne et al. 2006), some olfactory receptors have been built upon protein homology alone because of their restricted expression.
Locus and variant types were annotated according to established standards (Harrow et al. 2006), with the modification that, within the MHC region, the artefact locus has been used to tag historically annotated structures that are no longer deemed valid.
HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQA1, and HLA-DQB1 allele types were assessed by comparison against the IMGT/HLA database (http://www.ebi.ac.uk/imgt/hla/; Marsh et al. 2005).
Annotation status of haplotypes
The PGF, COX, and QBL haplotypes have already been annotated in detail (Stewart et al. 2004; Traherne et al. 2006). It was decided, however, to re-annotate and update this annotation to maintain consistency between all eight haplotypes with the current supporting evidence and pipeline analyses. The SSTO haplotype was manually annotated de novo. The new annotation from the PGF haplotype was projected through a DNA–DNA alignment to each of the remaining haplotypes (APD, DBB, MANN and MCF) where possible. This projection was checked thoroughly and non-alignable regions were manually adjusted (including the C4 and HLA-DRB1 hypervariable regions). Polyadenylation sites and signals were not annotated for haplotypes APD, DBB, MANN and MCF because of time constraints. In the main, however, these features may be assumed to correspond to the same positions as in the first four haplotypes.
Combination of variation and annotation data
By employing a series of Perl scripts, the array of haplotype variation was combined with the annotation of gene loci, repeat elements and microsatellites, extracted from the Vertebrate Genome Annotation (VEGA) database in general feature format (GFF; http://www.sanger.ac.uk/Software/formats/GFF/), to determine the variation status of all loci.
Distribution of sequenced HLA haplotypes in Europeans
To assess the distribution of sequenced haplotypes at the population level, 180 founder haplotypes were reconstructed using genotypic data from Centre d’Etude Polymorphisme Humain (CEPH) trios (de Bakker et al. 2006). A ~214 kb segment spanning the HLA–DRB1–DQB1 genes was selected for the analyses. This segment, represented by 54 SNPs, is delimited by rs2187823 and rs2856691, with NCBI build 36 chromosome 6 coordinates 32547486 and 32761413, respectively. Phased haplotypes with known HLA–DRB1–DQB1 alleles were then used to construct a neighbor-joining tree (Kumar et al. 2001) and a phylogenetic network (Bandelt et al. 1999).
Resources
All sequences presented in this paper have been submitted to the EMBL/GenBank/DNA Data Bank of Japan (DDBJ) database and allocated accession numbers. For clarity, all bacterial artificial chromosome (BAC) clones are referred to using their accession numbers. The annotation of each haplotype has been entered in the VEGA database and is accessible through its browser (http://www.VEGA.sanger.ac.uk). All variations from the study were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP) using the submitter handle SI_MHC_SNP.
BAC clones from the CHORI-501 (PGF) and CHORI-502 (COX) libraries can be requested from BACPAC resources (http://www.bacpac.chori.org/). Clones from the other libraries can be requested from john.elliott@ualberta.ca.
The web site for the MHC Haplotype Project provides links to various data resources (http://www.sanger.ac.uk/HGP/Chr6/MHC/).
DAS sources for all substitutions and indels are available from http://www.das.ensembl.org/das as follows:
-
ens_35_COX_SNP ens_35_COX_DIP
-
ens_35_QBL_SNP ens_35_QBL_DIP
-
ens_35_SSTO_SNP ens_35_SSTO_DIP
-
ens_35_APD_SNP ens_35_APD_DIP
-
ens_35_DBB_SNP ens_35_DBB_DIP
-
ens_35_MANN_SNP ens_35_MANN_DIP
-
ens_35_MCF_SNP ens_35_MCF_DIP
These can be accessed via the VEGA browser.
Results and discussion
Variation analysis
One of the main aims of the MHC Haplotype Project was to generate a comprehensive variation map of this most variable region of the human genome. To achieve this, eight haplotypes were sequenced and subjected to variation analysis. Table 1 details the lengths of the sequence contigs, the number of sequence gaps and the allelic types of major HLA loci for each haplotype. Of the eight haplotypes sequenced, three have already been described: PGF and Cox (Stewart et al. 2004) both of which formed single contigs of approximately 4.7 Mb, and QBL (Traherne et al. 2006), of approximately 4.2 Mb but with five gaps. The remaining haplotypes sequenced all contained gaps, their coverage ranging from 2.33 Mb (DBB with 28 gaps) to 4.19 Mb (MANN with 10 gaps).
For the variation analysis, each of the above haplotypes was compared with the PGF reference sequence, resulting in the identification of 44,544 variations (37,451 substitutions and 7,093 indels, Table 2), which have all been submitted to dbSNP. The success of this exercise is illustrated by the fact that examination of this public database (NCBI dbSNP build 127, March 2007) showed that there were only a further 19,598 variations, submitted by other laboratories, in this region which were not identified by this project. In accordance with the annotation that we also generated for each haplotype (see below), the variations shown in Table 2 were further classified as untranslated region (UTR), exonic, intronic, intergenic and eight more sub-categories (Table 3). Coding substitutions, which are of particular interest with respect to altered functionality, were further classified as synonymous, non-synonymous conservative, or non-synonymous non-conservative and grouped depending on whether they affected HLA or other genes (Table 4). The actual variations and affected amino acids can be viewed using the VEGA browser as illustrated in Fig. 1 and described in the corresponding section later on. In addition, we have analysed all haplotype sequences for inversions, which represent another important variation category that has been linked to genomic disorders (Shaw and Lupski 2004). Using Ssaha2 (Ning et al. 2001), we found no evidence of any inversion polymorphism within the generated sequences but could not exclude large-scale (e.g. involving entire MHC) inversions with breakpoints outside the MHC regions sequenced here.
Gene annotation
There have been several previous annotations of the gene content of the MHC (Horton et al. 2004; Mungall et al. 2003; Stewart et al. 2004; The MHC Sequencing Consortium 1999; Traherne et al. 2006). The maximum region annotated in this study extends from the telomeric ZNF452 gene in the MHC extended class I region (COX haplotype) to the centromeric ZBTB9 gene just telomeric of the MHC extended class II region (PGF and SSTO haplotypes). The PGF haplotype (Stewart et al. 2004) remains the longest complete MHC haplotype, encompassing 320 annotated loci with 1,267 variants. The number of variants ascribed to each locus-type is listed in Table 5. A comparison of the statistics for loci in each haplotype is shown in Table 6.
VEGA database and browser
The VEGA database provides access to gene annotation of the eight MHC haplotype sequences, a valuable public resource and a means of integrating annotation and variation data. The VEGA database also provides the facility to download nucleotide or peptide sequences for genes of interest, by selecting ‘export cDNA’ or ‘export peptide’ from the menu obtained by clicking on gene cartoons in the VEGA ‘detailed view’ or ‘basepair view’ window. From these, any desired alignments can be made. Variation data may be viewed in the browser linked to a distributed annotation system (DAS) source of any given variation (see “Materials and methods”). This is illustrated An example of the use of this browser to view a C to T substitution is illustrated for the OR2J1 locus (Fig. 1). An overview of the genomic environment is given in Fig. 1a, showing the gene within a cluster of olfactory gene loci on chromosome 6. The detailed view (Fig. 1b) shows OR2J1 with associated variations in all haplotypes. The basepair view (Fig. 1c) illustrates the presence of the C/T substitution in all haplotypes except MCF, and its positioning above the translated sequence, at the first position of a CAG codon, indicating the presence of a stop codon instead of glutamine.
Annotation changes
In addition to loci annotated in the previous studies, newly recognised with official Hugo Gene Nomenclature Committee (HGNC) symbols have also been annotated. These have included the mitochondrial coiled–coil domain protein 1 gene MCCD1 (Semple et al. 2003) and the related unprocessed pseudogenes MCCD1P1 and MCCD1P2, as well as the zinc-finger and BTB domain-containing protein gene ZBTB9, annotated at the very centromeric boundary of the sequenced region.
The C6orf21 gene (De Vet et al. 2003; XXbac–BPG32J3.17-001) of the MHC class III region was annotated as a separate locus from the adjacent centromeric locus LY6G6D (splice variants XXbac–BPG32J3.4-001 and XXbac–BPG32J3.4-002). There was, however, a further coding splice variant of LY6G6D (XXbac–BPG32J3.4-004), which spanned not only the other LY6G6D splice variants but also C6orf21, suggesting that this is a possible so-called chimeric transcript (Parra et al. 2006).
HLA-DRB1 hypervariable region
Of the five newly annotated MHC haplotypes, APD alone exhibited the HLA–DRBDR52 antigenic specificity found on DRB1*3, DRB1*05 (DRB1*11 and DRB1*012) and DR6 (DRB1*13 and DRB1*14) haplotypes and encoded by HLA–DRB3, whereas the remainder (SSTO, DBB, MANN and MCF) exhibited the DR53 specificity, encoded by HLA–DRB4, here annotated for the first time in genomic sequence. The HLA–DRB53 sequences included three known loci (HLA–DRB4, HLA–DRB7 and HLA–DRB8), as well as three novel pseudogenes (DASS–218M11.1, DASS–23B5.1 and DASS–23B5.2). DASS–23B5.1 corresponds to a pseudogene derived from the gene for the protein kinase, interferon-inducible double-stranded RNA dependent activator (Chida et al. 2001) for which the symbol PRKRAP1 has now been recognised. A further processed pseudogene, FAM8A5P (Jamain et al. 2001), was also annotated in the DR53 specificity.
HLA-V and HLA-P
Our analysis showed that the two unprocessed class I pseudogenes HLA-V and HLA-P ( previously HLA-75 and HLA-90, Geraghty et al. 1992) should in fact be merged together; individually they merely represented the 5′ and 3′ portions of a single unprocessed pseudogene, separated by repeat elements. According to our annotation guidelines (see “Materials and methods”), the newly merged locus was assigned the symbol from the 3′ component, in this case, HLA-P. Best-in-genome nucleotide evidence was found to support five transcript variants at the 5′ end, which, together with evidence for continued locus-transcription, led us to designate the locus as a transcribed pseudogene. Because transcription appears to still occur at this locus, it was, therefore, designated as a transcribed pseudogene. A further six expressed pseudogenes were identified in the MHC region (HLA–DPB2, HLA-J, CYP21A1P, HLA–DRB6, HLA–L and PPP1R2P1).
RCCX hypervariable region
This module within the MHC class III region, named for its gene content (RP-C4A/B-CYP21-TNXB), may be duplicated or triplicated (Chung et al. 2002), and the pseudogenes CYP21A1P, TNXA and STK19P contain the complement component gene, C4, in either or both of the two versions, C4A and C4B (Awdeh and Alper 1980). This gene may also be present in either long (C4AL, C4BL) or short (C4AS, C4BS) forms depending on the presence or absence of an inserted HERVC4 element in intron 9. Contrary to our previous annotation (Stewart et al. 2004) see also legend to (Fig. 2), the PGF haplotype now appears to possess an arrangement in which C4AL precedes C4BL, whereas COX has a single module with C4BS and QBL has a single module with C4AS (Traherne et al. 2006). For the new haplotype sequences reported in this paper, SSTO was bimodular with two copies of C4BL, whereas DBB was bimodular with C4AL followed by C4BS. Although a sequence gap was present in MCF, this haplotype appeared to be bimodular in that, although the telomeric copy of the C4 gene could not be identified, there was evidence for the pseudogenes CYP21A1P, TNXA and STK19P in a telomeric module. The second centromeric module in MCF contained C4AL. The RCCX region in the APD and MANN haplotypes was incomplete because of sequence gaps.
C6orf205
Variability in the C6orf205 gene has been reported to consist of extension of the minisatellite in exon 2 from 27 copies in PGF and COX to 31 copies in QBL (Traherne et al. 2006). In the newly annotated haplotypes, we found the minisatellite to extend to 29 in MANN. The APD, DBB and MCF possessed 27 copies. There was a sequence gap in this region in the SSTO haplotype.
MICA
The known allelic polymorphism of MICA reported for the DRB1*03 QBL cell line sequence, in which a four-base insertion (GCGT) extended the open reading frame in coding exon 5 haplotype (Traherne et al. 2006), was also present in the DRB1*07 MANN haplotype. The insertion was absent from PGF, COX and SSTO. No sequence was available in APD, DBB and MCF for this gene.
PPP1R2P1
The intronless pseudogene PPP1R2P1 reported to have a full-length open reading frame in the PGF, COX and QBL haplotypes (Stewart et al. 2004; Traherne et al. 2006) was found to have a similar open reading frame in the DBB and MANN haplotypes but to have the frameshift mutation seen in the original chromosome reference sequence (Mungall et al. 2003) in the SSTO, APD and MCF haplotypes.
PSORS1C1
The QBL haplotype remains the only one in which there was a single nucleotide deletion in a polyC tract of exon 5 (Traherne et al. 2006). DBB, MANN and MCF resembled PGF and COX. No sequence was available for this gene in SSTO or APD.
POU5F1
The PGF haplotype has been reported to have a disrupted start codon for alternative splice variant of POU5F1 (Traherne et al. 2006). This disruption was not present in COX or QBL nor was it present in the further haplotypes reported in this paper, namely SSTO, DBB, MANN and MCF. APD had no sequence in this region.
OR2J1
This olfactory receptor OR2J1 has been reported to have both functional and non-functional alleles (Ehlers et al. 2000), the latter the result of a premature stop codon at amino acid position 194 introduced by a substitution in the coding sequence. In our annotation, we found the PGF and MCF haplotypes to contain the full-coding sequence, whereas the COX, QBL SSTO, APD, DBB and MANN haplotypes to contain the truncated sequence as an unprocessed pseudogene (see above and Fig. 1).
Other annotation differences
Other loci included in the current but not the previous PGF annotation were HCG4P11, HCG4P8, HCG4P7, HCG4P5, HCG4P3 and the loci without symbols listed in Table 7. Previously annotated loci not annotated in this study or considered artefacts because they did not reach our current standards of annotation included HLA-X, C6orf215, HCG2P7, HCG8, HCP5P2, HCP5P3, HCP5P6, HCP5P12, HCP5P13, HCP5P14, HCP5P15, HCG8 and HCG26.
Non-canonical splice sites
Eight variants within six loci were shown to exhibit haplotypic variation at their splice sites (canonical to non-canonical motif; Table 8). These variations may affect the gene expression at the post-transcriptional level. Hoarau et al. (2004, 2005) have already described the differential splicing within the HLA–DQA1 locus, and this can clearly be seen by comparing the new HLA–DQA1 annotation through the VEGA genome browser.
Combination of variation and annotation data
The data for sequence contig length, gaps, variation rate within haplotypes and PGF coding gene annotation have been combined in the map in Fig. 2. This illustrates the concentration of variation around the HLA gene loci, specifically in 3 areas: around HLA-F, HLA-G and HLA-A; around HLA-C and HLA-B; and around HLA-DRB1, HLA-DQA1, HAL-DQB1, HLA-DQA2 and HLA-DQB2. The variation status of genes of the PGF haplotype is shown in Table 9.
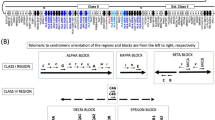
Variation and annotation map of eight MHC haplotypes. The map represents the complete reference sequence (orange bar split into three 1.6 Mb sections) labelled PGF and marked with a scale (Mb) and approximate megabase positions on the NCBI36 build of chromosome 6 (grey milestones). Below the reference sequence are arrows representing gene positions and orientations colour-coded for variation status (invariable, black; with synonymous variation only, green; with non-synonymous, conservative variation, red; with non-synonymous, non-conservative variation, purple; see Table 8) and their symbols on a band denoting MHC class (extended class I, green; class I, yellow; class III, pale orange; class II, light blue; extended class II, pink; outside MHC, pale grey). Above the reference sequence, coloured bands represent the sequences of the other seven haplotypes (COX, orange; QBL, mauve; APD, yellow; DBB, green; MANN, light blue; SSTO, dark blue; MCF, purple) with sequence gaps in dark grey; the RCCX hyper-variable region shown with green (C4A block) and/or red (C4B block) or black (block absent), and the HLA–DRB hyper-variable region in shades of blue-green. Above each haplotype bar, a bar-graph represents total variation between the haplotype and the reference sequence (total variations/10 kb) in dark red. Re-examination of the sequence AL645922 from the PGF haplotype, which contains the RCCX region, has shown that the original assembly was erroneous. Correction of these errors leads us now to the conclusion that the C4A gene precedes the C4B gene in this clone sequence. This new gene order is reflected in Fig. 2
As well as the variations reported above, major indels revealed as breaks in cross_match discrepancy lists and analysed by RepeatMasker are given in Table 10. Many of these have been previously reported (Dangel et al. 1994; Dunn et al. 2003; Dunn et al. 2002; Gaudieri et al. 1999; Horton et al. 1998; Kulski and Dunn 2005; Stewart et al. 2004). These indels were most frequently but not exclusively associated with AluY elements.
Four of these major indels were complex and designated as complex regions A, B, C and D in Table 10. They include three known regions from the comparison of the PGF and COX haplotypes (Stewart et al. 2004). Complex region A (involving MIR, MER41B, MER115, AluSx, Flam_C, AluSg, AluY, AluSx, L2 and MER38 elements) maps between TRIM27 and C6orf100 and was found to be deleted in COX but present in PGF, QBL, SSTO, MANN and MCF. Complex region B (involving L2 and AluY elements) maps to intron 1 of HLA–DRB1 and was also found by comparing PGF with COX, QBL, DBB, MANN and SSTO. Complex region C (SVA and low-complexity repeat elements) maps between HLA–DRB1 and HLA–DQA1 and was noted in COX as a deletion of the SVA and low-complexity repeats. Whereas, DBB, MANN and SSTO displayed the same deletion, as well as a telomeric deletion of AluSx/MIRb, QBL had both deletions plus that of an intervening 2.5 kb sequence containing Alu, L3 and MLT1A1 elements. Complex region D maps between HLA−DQA1 and HLA−DQB1 and is more complicated than previously reported. At the telomeric end, PGF lacks an L1PA4 fragment of >300 bp that is present in COX, QBL, SSTO and MCF and is also absent in DBB and MANN where it is interrupted by about 1.3 kb of SVA sequence. Centromeric to this PGF contains an AluSx, an AluY and an AluYd2, flanked by long interspersed nuclear element repeats, all deleted in the other haplotypes. Further towards the centromere there is an L1MA7 fragment, into which in PGF alone there are insertions of an AluSx followed by an AluY; a subsequent AluSg present in all haplotypes contains an insertion of 795 bp of SVA sequence in just COX and QBL. Finally, at the centromeric end of this region, PGF uniquely contains intact MER11C and LTR5 elements.
Representation of haplotypes within European populations
The eight haplotypes analysed in this study were selected on the basis of their association with type 1 diabetes and multiple sclerosis and their high population frequencies. To determine how representative these haplotypes are with respect to SNP haplotypic diversity in a population, we determined their distribution in the haplotypic tree space in the European population.
For this analysis, we selected a segment of ~214 kb, spanning the HLA–DRB1 and HLA–DQB1 genes in a population of European ancestry with known HLA allelic data (de Bakker et al. 2006). Phylogenetic analysis of 180 founder haplotypes derived from genotypic data (54 substitutions) shows that the eight haplotypes selected as part of the MHC Haplotype Project share identical HLA alleles over most of the tree space (Fig. 3a), representing almost the entire variation observed in the population assayed with the exception of two branches (DRB1*1103–DQB1*0301 and DRB1*0101–DQB1*0501).
Clusters of haplotypes in the European haplotypic diversity. Phylogenetic relationship of 180 founder SNP haplotypes from CEPH trios spanning a 214-kb segment of the MHC class II region, including the HLA-DRB1 and HLA-DQB1 genes (54 substitutions from rs2187823 to rs2856691). a Sequenced haplotypes are widely distributed in this NJ tree and represent the vast majority of the variation in the population sampled. Four-digit alleles are indicated for the corresponding DRB1 and DQB1 genes in each haplotype ID label to highlight the HLA haplotypic distribution based on the underlying nucleotide variation. The NJ tree was constructed using pairwise genetic distances considering the Kimura 2-parameters model without correction for rate variation among sites as implemented in the MEGA2 software (Kumar et al. 2001). b Each haplotype sequenced is associated to a single haplotype cluster. This phylogenetic network (Bandelt et al. 1999) also shows that clusters (shaded area) are constituted by one central haplotype and its derivatives. Circles represent individual haplotypes, and the size of the circle is proportional to the haplotype frequency. The length of the lines connecting nodes is relative to the distance between them, e.g. distances within shaded areas (clusters) never exceed three mutation steps. Cluster of haplotypes sharing HLA alleles with sequenced cell lines are named accordingly: COX and QBL: DRB1*0301 DQB1*0201–PGF: DRB1*1501 DQB1*0602–APD: DRB1*1301 DQB1*0603–MCF: DRB1*0401 DQB1*0301–DBB: DRB1*0701 DQB1*0303–SSTO: DRB1*0403 DQB1*0302–MANN: DRB1*0701 DQB1*0202. HLA haplotypes DRB1*1103–DQB1*0301 and DRB1*0101–DQB1*0501 indicate the two major haplotype clusters not represented in the MHC haplotype project data
Haplotype diversity in this sub-population is restricted to relatively few haplotype clusters (Fig. 3b). Each cluster consists of a founder haplotype, depicted by the most frequent and centrally located haplotype within the cluster. Recently derived haplotypes show lower frequencies and are connected to the central haplotype by relatively few mutation steps (in this case, up to three). This phylogenetic network clearly shows that all the sequenced haplotypes occupy central positions in their respective haplotypic groups. Inferences about phylogenetic relationships between haplotype clusters are, however, only approximate as a consequence of recombination events.
It should also be noted that SNP haploptypes derived from CEPH pedigrees of European ancestry by no means represent an exhaustive sampling of European diversity. Nevertheless, the sampling has been shown to represent the European population in the UK reasonably well (Ke et al. 2005). In conclusion, our analysis demonstrates that the HLA haplotypes selected for the MHC Haplotype Project are ancestral haplotypes, representative of MHC diversity in the European population.
Conclusion and outlook
The MHC Haplotype Project has succeeded in providing a new public resource for immune-linked disease and population genetic studies. First reports from studies using the resource indicate that it adds significant power to the identification and fine-mapping of disease-associated variations (Yeo et al. 2007). The data have also contributed to the recent identification of a first set of HLA tag SNPs, which hold great promise for future applications in clinical settings, e.g. to complement or replace classical HLA-typing in transplant medicine (de Bakker et al. 2006). While costs and other limitations of the current (capillary) sequencing technology have restricted our study to only few (eight) MHC haplotypes, the number of new variations found, combined with the fact that no variation plateau has yet been reached, indicates that there are many more variations to be discovered. The recent introduction of several new and massively parallel sequencing platforms (for review, see Bentley 2006) has created the opportunity to do just that by re-sequencing haplotypes and, eventually, entire genomes at the population level and as integral part of case control studies. Because of its wide-ranging medical importance, the MHC can be expected to be among the first regions of the human genome to be sequenced in this way. Such sequencing will provide the critical, and until now missing, data to identify causal variations and their underlying mechanisms on an unprecedented scale.
References
Allcock RJ, Atrazhev AM, Beck S, de Jong PJ, Elliott JF, Forbes S, Halls K, Horton R, Osoegawa K, Rogers J, Sawcer S, Todd JA, Trowsdale J, Wang Y, Williams S (2002) The MHC haplotype project: a resource for HLA-linked association studies. Tissue Antigens 59:520–521
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Awdeh ZL, Alper CA (1980) Inherited structural polymorphism of the fourth component of human complement. Proc Natl Acad Sci U S A 77:3576–3580
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Benson G (1999) Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580
Bentley DR (2006) Whole-genome re-sequencing. Curr Opin Genet Dev 16:545–552
Birney E, Clamp M, Durbin R (2004) GeneWise and Genomewise. Genome Res 14:988–995
Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268:78–94
Chida S, Hohjoh H, Hirai M, Tokunaga K (2001) Haplotype-specific sequence encoding the protein kinase, interferon-inducible double-stranded RNA-dependent activator in the human leukocyte antigen class II region. Immunogenetics 52:186–194
Chung EK, Yang Y, Rennebohm RM, Lokki ML, Higgins GC, Jones KN, Zhou B, Blanchong CA, Yu CY (2002) Genetic sophistication of human complement components C4A and C4B and RP-C4-CYP21-TNX (RCCX) modules in the major histocompatibility complex. Am J Hum Genet 71:823–837
Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu LC, Yu CY (1994) The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics 40:425–436
de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, Morrison J, Richardson A, Walsh EC, Gao X, Galver L, Hart J, Hafler DA, Pericak-Vance M, Todd JA, Daly MJ, Trowsdale J, Wijmenga C, Vyse TJ, Beck S, Murray SS, Carrington M, Gregory S, Deloukas P, Rioux JD (2006) A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38:1166–1172
De Vet EC, Aguado B, Campbell RD (2003) Adaptor signalling proteins Grb2 and Grb7 are recruited by human G6f, a novel member of the immunoglobulin superfamily encoded in the MHC. Biochem J 375:207–213
Down TA, Hubbard TJ (2002) Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res 12:458–461
Dunn DS, Naruse T, Inoko H, Kulski JK (2002) The association between HLA-A alleles and young Alu dimorphisms near the HLA-J, -H, and -F genes in workshop cell lines and Japanese and Australian populations. J Mol Evol 55:718–726
Dunn DS, Inoko H, Kulski JK (2003) Dimorphic Alu element located between the TFIIH and CDSN genes within the major histocompatibility complex. Electrophoresis 24:2740–2748
Ehlers A, Beck S, Forbes SA, Trowsdale J, Volz A, Younger R, Ziegler A (2000) MHC-linked olfactory receptor loci exhibit polymorphism and contribute to extended HLA/OR-haplotypes. Genome Res 10:1968–1978
Gaudieri S, Kulski JK, Dawkins RL, Gojobori T (1999) Different evolutionary histories in two subgenomic regions of the major histocompatibility complex. Genome Res 9:541–549
Geraghty DE, Koller BH, Hansen JA, Orr HT (1992) The HLA class I gene family includes at least six genes and twelve pseudogenes and gene fragments. J Immunol 149:1934–1946
Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C, Hubbard T, Antonarakis SE, Guigo R (2006) GENCODE: producing a reference annotation for ENCODE. Genome Biol 7(Suppl 1):1–9
Hoarau JJ, Cesari M, Caillens H, Cadet F, Pabion M (2004) HLA DQA1 genes generate multiple transcripts by alternative splicing and polyadenylation of the 3′ untranslated region. Tissue Antigens 63:58–71
Hoarau JJ, Festy F, Cesari M, Pabion M (2005) A new splicing acceptor site and poly(A) +sequence signal within DQA1*0401 and DQA1*0501 mRNA 3¢UTR contribute to increase the extraordinary diversity of mRNA isoforms. Immunogenetics 57:182–188
Horton R, Niblett D, Milne S, Palmer S, Tubby B, Trowsdale J, Beck S (1998) Large-scale sequence comparisons reveal unusually high levels of variation in the HLA-DQB1 locus in the class II region of the human MHC. J Mol Biol 282:71–97
Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S (2004) Gene map of the extended human MHC. Nat Rev Genet 5:889–899
Jamain S, Girondot M, Leroy P, Clergue M, Quach H, Fellous M, Bourgeron T (2001) Transduction of the human gene FAM8A1 by endogenous retrovirus during primate evolution. Genomics 78:38–45
Ke X, Miretti MM, Broxholme J, Hunt S, Beck S, Bentley DR, Deloukas P, Cardon LR (2005) A comparison of tagging methods and their tagging space. Hum Mol Genet 14:2757–2767
Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, Browne P, Castro M, Cochrane G, Duggan K, Eberhardt R, Faruque N, Hoad G, Kanz C, Lee C, Leinonen R, Lin Q, Lombard V, Lopez R, Lorenc D, McWilliam H, Mukherjee G, Nardone F, Pastor MP, Plaister S, Sobhany S, Stoehr P, Vaughan R, Wu D, Zhu W, Apweiler R (2007) EMBL nucleotide sequence database in 2006. Nucleic Acids Res 35:D16–D20
Kulski JK, Dunn DS (2005) Polymorphic Alu insertions within the major histocompatibility complex class I genomic region: a brief review. Cytogenet Genome Res 110:193–202
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17(12):1244–1245
Lechler R, Warrens A (2000) HLA in health and disease. Academic, 2000
Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, Mayr WR, Parham P, Petersdorf EW, Sasazuki T, Schreuder GM, Strominger JL, Svejgaard A, Terasaki PI, Trowsdale J (2005) Nomenclature for factors of the HLA system, 2004. Hum Immunol 66:571–636
Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, Morrison J, Whittaker P, Lander ES, Cardon LR, Bentley DR, Rioux JD, Beck S, Deloukas P (2005) A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am J Hum Genet 76:634–646
Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, Gilbert JG, Clamp ME, Bethel G, Milne S, Ainscough R, Almeida JP, Ambrose KD, Andrews TD, Ashwell RI, Babbage AK, Bagguley CL, Bailey J, Banerjee R, Barker DJ, Barlow KF, Bates K, Beare DM, Beasley H, Beasley O, Bird CP, Blakey S, Bray-Allen S, Brook J, Brown AJ, Brown JY, Burford DC, Burrill W, Burton J, Carder C, Carter NP, Chapman JC, Clark SY, Clark G, Clee CM, Clegg S, Cobley V, Collier RE, Collins JE, Colman LK, Corby NR, Coville GJ, Culley KM, Dhami P, Davies J, Dunn M, Earthrowl ME, Ellington AE, Evans KA, Faulkner L, Francis MD, Frankish A, Frankland J, French L, Garner P, Garnett J, Ghori MJ, Gilby LM, Gillson CJ, Glithero RJ, Grafham DV, Grant M, Gribble S, Griffiths C, Griffiths M, Hall R, Halls KS, Hammond S, Harley JL, Hart EA, Heath PD, Heathcott R, Holmes SJ, Howden PJ, Howe KL, Howell GR, Huckle E, Humphray SJ, Humphries MD, Hunt AR, Johnson CM, Joy AA, Kay M, Keenan SJ, Kimberley AM, King A, Laird GK, Langford C, Lawlor S, Leongamornlert DA, Leversha M et al (2003) The DNA sequence and analysis of human chromosome 6. Nature 425:805–811
Ning Z, Cox AJ, Mullikin JC (2001) SSAHA: a fast search method for large DNA databases. Genome Res 11:1725–1729
Parra G, Reymond A, Dabbouseh N, Dermitzakis ET, Castelo R, Thomson TM, Antonarakis SE, Guigo R (2006) Tandem chimerism as a means to increase protein complexity in the human genome. Genome Res 16:37–44
Salamov AA, Solovyev VV (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res 10:516–522
Searle SM, Gilbert J, Iyer V, Clamp M (2004) The otter annotation system. Genome Res 14:963–970
Semple JI, Ribas G, Hillyard G, Brown SE, Sanderson CM, Campbell RD (2003) A novel gene encoding a coiled-coil mitochondrial protein located at the telomeric end of the human MHC Class III region. Gene 314:41–54
Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum Mol Genet Special Issue 13(1):R57–R64
Shiina T, Ota M, Shimizu S, Katsuyama Y, Hashimoto N, Takasu M, Anzai T, Kulski JK, Kikkawa E, Naruse T, Kimura N, Yanagiya K, Watanabe A, Hosomichi K, Kohara S, Iwamoto C, Umehara Y, Meyer A, Wanner V, Sano K, Macquin C, Ikeo K, Tokunaga K, Gojobori T, Inoko H, Bahram S (2006) Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 173:1555–1570
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147:195–197
Smith WP, Vu Q, Li SS, Hansen JA, Zhao LP, Geraghty DE (2006) Toward understanding MHC disease associations: partial resequencing of 46 distinct HLA haplotypes. Genomics 87:561–571
Stewart CA, Horton R, Allcock RJ, Ashurst JL, Atrazhev AM, Coggill P, Dunham I, Forbes S, Halls K, Howson JM, Humphray SJ, Hunt S, Mungall AJ, Osoegawa K, Palmer S, Roberts AN, Rogers J, Sims S, Wang Y, Wilming LG, Elliott JF, de Jong PJ, Sawcer S, Todd JA, Trowsdale J, Beck S (2004) Complete MHC haplotype sequencing for common disease gene mapping. Genome Res 14:1176–1187
The MHC Sequencing Consortium (1999) Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 401:921–923
The Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447:661–678
Traherne JA, Horton R, Roberts AN, Miretti MM, Hurles ME, Stewart CA, Ashurst JL, Atrazhev AM, Coggill P, Palmer S, Almeida J, Sims S, Wilming LG, Rogers J, de Jong PJ, Carrington M, Elliott JF, Sawcer S, Todd JA, Trowsdale J, Beck S (2006) Genetic analysis of completely sequenced disease-associated MHC haplotypes identifies shuffling of segments in recent human history. PLoS Genet 2:e9
Yeo TW, De Jager PL, Gregory SG, Barcellos LF, Walton A, Goris A, Fenoglio C, Ban M, Taylor CJ, Goodman RS, Walsh E, Wolfish CS, Horton R, Traherne J, Beck S, Trowsdale J, Caillier SJ, Ivinson AJ, Green T, Pobywajlo S, Lander ES, Pericak-Vance MA, Haines JL, Daly MJ, Oksenberg JR, Hauser SL, Compston A, Hafler DA, Rioux JD, Sawcer S (2007) A second major histocompatibility complex susceptibility locus for multiple sclerosis. Ann Neurol 61:228–236
Acknowledgements
The authors thank C.-K. Chen and all the staff of the DNA Sequencing Division and the HAVANA group (http://www.sanger.ac.uk/HGP/havana) at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk) for their assistance. This work was supported by a joint grant (048880) from the Wellcome Trust to J.A.T., J.T., S.B. and S.S., and a Wellcome Trust/Juvenile Diabetes Research Foundation grant to J.A.T.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Horton and Gibson contributed equally to this work.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Horton, R., Gibson, R., Coggill, P. et al. Variation analysis and gene annotation of eight MHC haplotypes: The MHC Haplotype Project. Immunogenetics 60, 1–18 (2008). https://doi.org/10.1007/s00251-007-0262-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00251-007-0262-2