Abstract
Engineering biosynthetic pathways in heterologous microbial host organisms offers an elegant approach to pathway elucidation via the incorporation of putative biosynthetic enzymes and characterization of resulting novel metabolites. Our previous work in Escherichia coli demonstrated the feasibility of a facile modular approach to engineering the production of labdane-related diterpene (20 carbon) natural products. However, yield was limited (<0.1 mg/L), presumably due to reliance on endogenous production of the isoprenoid precursors dimethylallyl diphosphate and isopentenyl diphosphate. Here, we report incorporation of either a heterologous mevalonate pathway (MEV) or enhancement of the endogenous methyl erythritol phosphate pathway (MEP) with our modular metabolic engineering system. With MEP pathway enhancement, it was found that pyruvate supplementation of rich media and simultaneous overexpression of three genes (idi, dxs, and dxr) resulted in the greatest increase in diterpene yield, indicating distributed metabolic control within this pathway. Incorporation of a heterologous MEV pathway in bioreactor grown cultures resulted in significantly higher yields than MEP pathway enhancement. We have established suitable growth conditions for diterpene production levels ranging from 10 to >100 mg/L of E. coli culture. These amounts are sufficient for nuclear magnetic resonance analyses, enabling characterization of enzymatic products and hence, pathway elucidation. Furthermore, these results represent an up to >1,000-fold improvement in diterpene production from our facile, modular platform, with MEP pathway enhancement offering a cost effective alternative with reasonable yield. Finally, we reiterate here that this modular approach is expandable and should be easily adaptable to the production of any terpenoid natural product.
Similar content being viewed by others
Introduction
Metabolic engineering to introduce heterologous biosynthetic pathways into microbial host organisms offers an elegant method for not only the production of “natural” products, but also for pathway elucidation and biochemical enzymatic investigation. In vivo production of appropriate precursors as substrates for putative, recombinantly coexpressed biosynthetic enzymes, coupled with characterization of the resulting product, provides a means for investigating enzymatic specificity and hence, their role in metabolic pathways. Emerging genomic sequences from a variety of organisms highlights the need to functionally investigate the encoded multifaceted metabolism. Central to any metabolic engineering effort is the ability to redirect appreciable carbon flux from central metabolism toward desired heterologous end products, which assumes particular importance when the production of novel metabolites necessitates their structural analysis.
Among the most biochemically interesting biosynthetic pathways are those producing natural products, which present a rich source of chemical diversity. However, these compounds are often found in low abundance, are difficult to isolate, and are refractory to chemical synthesis, thus making them a challenge for investigation (Clardy and Walsh 2004). Among the natural products, over 100,000 low molecular weight compounds known as secondary metabolites have been identified. Although, by definition, these compounds are not required for normal growth and development, they do perform a variety of biological functions, including antimicrobial defense and growth regulation, providing the selective force for such biochemical evolution (Firn and Jones 2003). In addition, about half of all prescribed therapeutic drugs in use today, particularly the invaluable antibiotics, have their origins in natural products, either directly or as semisynthetic derivatives (Newman and Cragg 2007).
The largest family of natural products is the terpenoids or isoprenoids (>40,000), comprised of various numbers of five carbon isoprene units, which are derived from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) precursors (McGarvey and Croteau 1995). In nature, there are two routes leading to IPP and DMAPP, the methyl erythritol phosphate (MEP) and mevalonic acid (MEV) pathways (Scheme 1). While the MEV pathway, found primarily in eukaryotes, begins with acetyl-CoA, the prokaryotic and plant plastid MEP pathway is initiated by condensation of glyceraldehyde-3-phosphate and pyruvate (Rohmer 2008). The resulting IPP and in the case of the MEP pathway, its allylic isomer DMAPP, can be interconverted to balance their use in production of longer chain isoprenoid precursors. These are produced by prenyltransferases via the addition of IPP to allylic diphosphate precursors, beginning with DMAPP, to sequentially produce geranyl diphosphate, farnesyl diphosphate, and geranylgeranyl diphosphate (GGPP), which are the immediate precursors of the C10 mono-, C15 sesqui-, and C20 diterpenoids, respectively.
a MEP pathway found in plant plastids and prokaryotes. DXS: DXP synthase, DXR: DXP reductase, IspD: ME-4-phosphate cytidyltransferase, IspE: 4-(cyt-5′-diphospho)-ME kinase, IspF: ME-2,4-cyclodiphosphate synthase, IspG: HMB-4-diphosphate synthase, IspH: HMB-4-diphosphate reductase. b MEV pathway found in eukaryotes. AAS: acetoacteyl-CoA synthase, HMGS: HMG-CoA synthase, HMGR: HMG-CoA reductase, MK: mevalonate kinase, PMK: phosphomevalonate kinase, PMD: mevalonate diphosphate decarboxylase, IDI: IPP isomerase
We have previously reported development of a modular metabolic engineering system for facile investigation of diterpene synthases (Cyr et al. 2007). In particular, coexpression of a GGPP synthase with various combinations of the class II cyclases and class I synthases responsible for the two-step enzymatic conversion of GGPP into multicyclic labdane-related diterpenes, although this system can be used with any diterpene synthase (and with minor modification, any terpene synthase). However, the resulting diterpene yields were relatively low (<100 µg/L of recombinant bacterial culture), hindering use of the system for characterization of novel enzymatic activities. These initial investigations relied upon the endogenous pools of IPP and DMAPP. Previous work, generally targeted at production of individual or small groups of terpenoids, often carotenoids (C40 tetraterpenoids whose phytoene precursor is formed by condensation of two GGPP), has demonstrated that engineering the MEV and MEP pathways can dramatically increase isoprenoid yield (Das et al. 2007). Here, we report that such precursor pathway engineering also dramatically increases yield (up to >1,000-fold) in the context of our modular system, which provides a means for its use in de novo investigation of novel enzymatic activity and hence, corresponding metabolic pathways. Our investigations further highlight a number of practical issues that can be important considerations in the application of isoprenoid precursor pathway engineering.
Results
Initial optimization of growth conditions
To minimize the number of manipulations necessary to measure diterpene production, the bifunctional diterpene cyclase abietadiene synthase from Abies grandis (AgAS), which has both class II and I activities, was coexpressed with a GGPP synthase, also from A. grandis (AgGGPS), using our previously described pGGAS dual expression vector (Cyr et al. 2007). Transformation of this vector is sufficient to endow the resulting recombinant Escherichia coli with the ability to produce the expected abietadiene (Scheme 2). Production of this diterpene was analyzed under a variety of growth conditions, particularly variation in liquid growth media and length of postinduction culture fermentation. With this minimally engineered strain (pGGAS only), it was found that rich TB media ultimately enabled higher cell densities and diterpene yield compared to NZY media (Supplemental Fig. 1). This increase in yield was particularly evident with longer fermentations in TB media, which exhibited continued accumulation of abietadiene up to the point at which cell densities started to decrease (Fig. 1), as measured by absorbance at 600 nm (A600). Culture viability and hence, diterpene yield, was extended by growing the cells in liquid media alone, rather than the previously described 2% (weight per volume) hydrophobic bead containing mixed phase conditions (Supplemental Fig. 1). Perhaps due to its ability to sequester diterpene extracellularly, rather than the presumed accumulation in cell membranes, mixed phase cultures did result in more efficient production of diterpene compared to liquid media cultures, as evidenced by greater yields normalized for cell densities at the peak of cell growth; however, the usage of two-phase cultures resulted in poorer cell growth and lower net diterpene production. The results of these various preliminary experiments suggested a correlation between diterpene yield and cell density. No loss of diterpene, either from exogenously added controls or metabolically engineered production, was observed even after prolonged growths of up to a week in duration, suggesting that at the optimal 16 °C production growth temperature loss to volatilization or bacterial degradation is minimal (Supplemental Fig. 2). Through these various improvements in growth conditions, abietadiene yield was increased to over 2 mg/L bacterial culture, representing a significant improvement in net yield from the previously reported ≤0.1 mg/L (Cyr et al. 2007).
Engineering the endogenous MEP pathway
Previously reported engineering of the endogenous MEP isoprenoid precursor pathway indicates that terpenoid yield in E. coli can be increased by overexpression of three of the relevant enzymatic genes; 1-deoxy-D-xylulose-5-phosphate synthase (dxs), 1-deoxy-D-xylulose-5-phosphate reductase (dxr), and/or isopentenyl diphosphate isomerase (idi) (Das et al. 2007). However, these studies differed in which genes were found to be beneficial and overexpressed, at most, only two of these three. Thus, the ability of overexpression of these MEP pathway genes to increase diterpene production was explored here by the creation of compatible expression vectors carrying either single genes (pDXS, pDXR, pIDI), two gene combinations (pSR, pIS, pIR), or all three genes (pIRS). Each of these was cotransformed with pGGAS and abietadiene yield from the resulting recombinant E. coli determined in duplicate (Table 1). Overexpression of idi, either alone or in combination with dxs and/or dxr, consistently increased diterpene yield. By contrast, overexpression of dxr, dxs, or both actually decreased diterpene yield. In addition, while coexpression of idi with either dxs or dxr increased diterpene yield relative to overexpression of dxs or dxr alone, the resulting level remained below that of the control (pGGAS only) strain. Nevertheless, overexpression of dxs, dxr, and idi together increased diterpene yield, even beyond that observed with idi alone, although this was limited to an approximate threefold increase relative to the control. To summarize, in three independent trial series, the order of increasing effectiveness in diterpene production for the MEP pathway combinations was consistently found to follow the corresponding trend:
Addition of a heterologous MEV pathway
Dramatic increases in bacterial isoprenoid yield resulting from the introduction of a heterologous MEV pathway from Sacchromyces cereviseae have been reported by the Keasling group (Martin et al. 2003). This was accomplished by providing the enzymatic genes necessary for the production of mevalonate from acetyl-CoA (in a synthetic operon encoding the “top half” of the MEV pathway) with the pMevT vector, while those for the production of IPP from mevalonate (in a synthetic operon encoding the “bottom half” of the MEV pathway), along with idi, were provided by the pMBI vector. Notably, feeding mevalonate to recombinant strains containing pMBI only was sufficient to essentially replicate the increased isoprenoid yield obtained by addition of the full MEV pathway (i.e., use of both pMevT and pMBI). By design, our modular metabolic engineering system is compatible with pMBI, which has been previously used to increase diterpene yield (Morrone et al. 2008). However, due to the associated expense of mevalonate feeding, incorporation of the full MEV pathway also was pursued.
In an attempt to reduce the number of plasmids necessary for diterpene production, AgGGPS was transferred from pGGAS to the synthetic operon in pMBI, producing pMBID. Unfortunately, coexpression of pMBID with AgAS alone (from a separate pET-based expression vector rather than pGGAS) resulted in the production of very little abietadiene. This may be due to the noticeably poor growth of the corresponding recombinant strain, for which cell densities in shake flask cultures were A600 ≤ 4.0.
The Keasling group has previously reported that incorporation of an additional copy of a truncated form of 3-hydroxy-3-methylglutaryl-CoA reductase (hmgr) to the “bottom half” of the MEV pathway further increases isoprenoid yield (Pitera et al. 2007). Further, pMevT is not compatible with our modular metabolic engineering system. In particular, while pMevT is compatible with pGGAS, it is incompatible with the pDEST vector series typically used for expression of the class I diterpene synthases catalyzing the secondary rearrangement and/or cyclization reaction in labdane-related diterpene biosynthesis (Cyr et al. 2007). Hence, the synthetic operon from pMevT was moved into a dual expression vector that also encoded an additional copy of the truncated hmgr gene under control of a separate promoter, creating pMTH. Coexpression of pMBI with pGGAS, along with feeding mevalonate to 20 mM, increased the yield of abietadiene. Mevalonate feeding experiments determined that amounts of 10 mM were most efficiently converted to diterpene; while 20 mM provided the highest net yield, it was not as efficiently converted to diterpene as 10 mM concentrations (Table 1). Additionally, it was found that amounts of 40 mM mevalonate appear toxic to E. coli, as evidenced by a complete inhibition of cell growth after mevalonate addition at induction and minimal diterpene yield. These results are similar to those reported earlier (Martin et al. 2003). Addition of the full MEV pathway using pMevT resulted in high abietadiene yield, which could be further increased by incorporation of the extra copy of truncated hmgr via use of pMTH instead of pMevT (Table 1).
Reoptimization of growth conditions
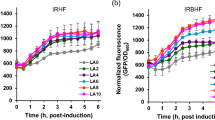
Given the significant increases seen with basic optimization of the growth conditions for the minimally engineered pGGAS contain strain of E. coli, more investigations were undertaken with the further engineered MEP and MEV isoprenoid precursor pathway strains. Initially, a variety of media conditions were tested to determine a suitable media recipe for the metabolic engineering of E. coli or diterpene production. Media recipes for NZY, buffered NZY (NZY B), buffered NZY + glycerol (NZY BG), TB, and buffered 2xNZY + 2% glycerol (FL) were tested. The richer media of TB and FL resulted in increased culture density and diterpene production (Fig. 2a). However, it was consistently found that glycerol significantly retarded initial growth. As a result, glycerol supplementation was initiated at induction. Additionally, the optimal glycerol concentration was determined to be 2% based on diterpene production and culture density in a series of biofermentations wherein glycerol content was varied within TB media (Fig. 2b). Growths relying on lactose autoinduction failed to produce as much diterpene product as did IPTG induction. Furthermore, the use of baffled flasks greatly enhanced culture density, presumably because of increased gas exchange. Baffled flasks shaken at 200 rpm with optimal media conditions routinely allowed culture densities with A600 >20. As suggested earlier, the increased cell density was correlated with an increase in diterpene production (Fig. 3) and coupled with the limitations of mevalonalactone, presumably due to toxicity, high density titers are desired. Finally, to supplement MEP pathway utilization, addition of pyruvate as a precursor was tested. Addition of 50 mM pyruvate to MEP pathway enhanced (pIRS) cultures resulted in an approximately twofold increase in diterpene production to a level of ∼14 mg/L (Fig. 4). The combination of the optimization of media, culturing conditions, and precursor feeding for abietadiene biofermentation production with the previously determined high yielding vector combinations from the MEP and MEV pathways given in Table 1 are depicted in Fig. 5.
Reoptimization of growth and media conditions tested using the minimally engineered MEV strain (pMBI + pGGAS) for abietadiene production in E. coli, with both net and normalized yields depicted. Mevalonate (20 mM) was added to shake flask cultures grown for 72 h in duplicate. a The results of abietadiene yield for the various media tested: NZY, NZY B, NZY BG, TB, and FL. b The results of abietadiene yield for the various amounts of glycerol (percentage v/v) applied to TB media. All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
Correlation of abietadiene production with cell density. Baffled flasks were utilized to grow cultures in TB media to final cell density averages of 15.2, 20.1, and 22.6. Extraction and analysis was performed 48 h after induction, at cell density peak. All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
Effects of pyruvate supplementation on abietadiene production from MEP pathway-engineered strains. Pyruvate supplementation added to media at the indicated concentrations was carried out in shake flask cultures grown in FL media for 72 h. a Net yield at the various pyruvate concentrations. b Normalized yield. All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
Optimal cell growth and production of abietadiene in metabolically engineered E. coli as a function of time. Biofermentation with pMBI + pGGAS supplemented with 20 mM mevalonolactone (triangles), pIRS + pGGAS (circles), and pMTH + pMBI + pGGAS (squares). Baffled shake flasks were used along with TB media optimized from previous experimental findings. Trials were performed in duplicate. a Cell growth over time. b Yield over time. c Normalized net yields. All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
Combining MEP and MEV pathway engineering
The increases in diterpene production observed through metabolic engineering by using the MEP and heterologous MEV pathway modules prompted us to test combinations using both pathways and incorporating multiple copies of the class II and class I modules. These combinations and multiplicities are listed in Table 2. Essentially, none of the attempts to incorporate both pathways or multiple copies of various biosynthetic enzymes resulted in higher yields compared to the heterologous MEV pathway alone. Growth was generally poor in the cultures requiring four modules and four markers. When yields were normalized for cell density, the combination of the MEP and MEV pathway with the minimally engineered strain (pMBI + pIRS + pGGAS) did provide amounts of abietadiene similar to the MEV strain just using pMBI (5.8 vs 5.6 mg/L/A600); however, the net yield of the combination strain was significantly lower. These results are consistent with the suggestion that only catalytic amounts of the enzymes are required, and rather, it is flux that is desired for high yield. Central to this appears to be the ability of the expressing culture as a whole to either have sufficient uptake of exogenous precursors or to perhaps produce a large precursor pool if using an endogenous supply; in either case, it suggests an advantage in net yield from high density cultures.
Bioreactor yields
Previous reports have suggested that the use of bioreactors enables higher cell density and greater isoprenoid yields (Newman et al. 2006). Thus, the ability of bioreactor grown cultures to further increase diterpene production in the context of our modular metabolic engineering system in E. coli was investigated. Previous work with our bioreactor (New Brunswick BioFlo110) demonstrated the effectiveness of pulsed feeding of glycerol and mevalonate for pMBI to 4% and 20 mM, respectively (Morrone et al. 2008). Briefly, every 12 h after induction up to 48 h, a solution of glycerol and mevalonate was injected, with the sum of the injections totaling up to the targeted 4% and 20 mM concentrations. In bioreactor growths under optimal media and growth conditions, we were able to obtain cell densities of A600 ≈15, with yields of ∼30 mg/L and ∼200 mg/L abietadiene for strains carrying pGGAS and pMBI, with either pMTH or mevalonate feeding, respectively (Fig. 6).
For comparison, the use of a bioreactor to increase yield from a strain with an engineered endogenous MEP pathway (i.e., the recombinant strain carrying the optimal pIRS + pGGAS) also was investigated. This led to a yield of ∼20 mg/L of abietadiene. Notably, while the diterpene yields are less for the MEP pathway compared to the MEV, it is important to distinguish that there is no need for either costly mevalonate supplementation or for the utilization of two plasmids (pIRS versus pMBI and pMTH). In addition, while use of a bioreactor did increase yield (quite dramatically in the case of pMBI coexpression with mevalonate supplementation), the downside is the expense of such instrumentation and their complexity relative to shake flask growths. To determine the yield from a relatively simple and cost effective approach, a recombinant strain carrying pIRS + pGGAS was grown in 1 L of FL media in a 2.8-L baffled flask, with 50 mM pyruvate supplementation, resulting in ∼16 mg of abietadiene, an amount that is sufficient for chemical characterization by nuclear magnetic resonance (NMR), as well as further biochemical studies (e.g., precursor/substrate feeding experiments).
Extension to other labdane-related diterpenes
Among the labdane-related diterpenes, there are three commonly found stereoisomers. These are designated by comparison to the A/B ring configuration found in abietadiene, which is then termed normal, with ent- and syn- stereoisomers also commonly found. We previously demonstrated use of our modular system for facile production of labdane-related diterpenes of all three configurations (Cyr et al. 2007). This was accomplished via dual expression vectors expressing AgGGPS and a stereospecific class II diterpene cyclase, forming a small series of pGGxC vectors wherein pGGnC, pGGeC, and pGGsC enable production of normal, ent-, or syn- copalyl diphosphate (CPP), respectively. These CPP intermediates, in turn, can then be reacted upon by class I stereospecific labdane-related diterpene synthases. To demonstrate that the optimization described above for production of abietadiene is more broadly applicable, we tested pGGeC and pGGsC, with coexpression of a relevant class I labdane-related diterpene synthase (i.e., pumpkin (Cucurbita maxima) ent-kaurene synthase (CmKS) or a rice (Oryzae sativa) syn-stemodene synthase (OsKSL11), respectively (Scheme 3)), under optimized conditions (i.e., in TB media with pMBI and pulsed feeding of 20 mM mevalonate). Yields in both cases were significantly higher than that previously reported (Cyr et al. 2007), even in shake flask cultures (Fig. 7), with further significant increases seen in bioreactor fermentations, much as observed for abietadiene production.
Comparison of diterpene yield from various CPS. Production of syn and ent diterpenes by metabolically engineered E. coli using the various class II cyclases depicted with minimal MEV pathway (i.e., pMBI + 20 mM mevalonate feeding). Biofermentation was performed in shake flask cultures with TB media and harvested 72 h postinduction in parallel duplicate trials. a Net values of diterpene produced. b Values of diterpene produced normalized for cell density. Error bars depict standard deviation (a) and propagated standard deviation, without covariance (b). All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
However, with both pGGeC and pGGsC, the net diterpene yields were lower than those observed for pGGAS. While the syn-CPP synthase in pGGsC is the only such enzyme known (Xu et al. 2004), there are many ent-CPP synthases known beyond that from Arabidopsis thaliana (AtCPS) utilized in the original version of pGGeC. Thus, we constructed variants of pGGeC using different ent-CPP synthases, specifically that from corn (Zea maize, An2 (Harris et al. 2005)), from rice (Oryza sativa, OsCPS2 (Prisic et al. 2004)), and the bacteria Bradyrhizobium japonicum (BjCPS; Morrone et al. 2009). With the exception of that constructed with BjCPS, the resulting variously derived pGGeC vectors allowed for sufficient culture growth with cell densities of similar values to those observed with abietadiene production (Supplemental Fig. 3). Notably, the version of pGGeC carrying An2 improved ent-kaurene yield threefold relative to the original AtCPS-derived construct (Fig. 7) and was found to offer higher yields with several different ent-kaurene synthases, indicating that the production of ent-CPP had been a limiting step (Supplemental Fig. 4).
Given the role for Mg2+ as an enzymatic cofactor for both classes of diterpene synthases, we investigated the ability of exogenously added Mg2+ to further increase yield (Fig. 8). While this did not improve yields of abietadiene, the addition of low millimolar concentrations (∼5 mM) of Mg2+ to FL media does increase diterpene yield with pGGsC and pGGeC(An2), although not pGGeC(AtCPS).
Effect of Mg2+ on diterpene yield. MEV pathway combinations consisting of pMBI + pGGxC + pDEST class I were assayed for diterpene yield under varying amounts of Mg2+ in FL media shake cultures with 20 mM mevalonate added and 72 h of cultivation. The various class II cyclases used in the pGGxC modules are indicated on the x-axis and grouped. Mg2+ concentrations (0 to 50 mM) are indicated above bars. Abietadiene synthase, kaurene synthase, and stemodene synthase were utilized in the pDEST class I modules with the appropriate pGGxC base vector. a Net diterpene yield. b Normalized yield. All data is from cultures grown in parallel, with error bars depicting the standard deviation of duplicate cultures
Discussion
The potential of metabolic engineering and synthetic biology is still being explored. For example, innovative work by the Keasling group has recently demonstrated that it is possible to metabolically engineer E. coli to produce large quantities of artemisinic acid, a usable precursor for semisynthetic production of the antimalarial sesquiterpenoid natural product and medically relevant drug artemisinin (Chang et al. 2007). This, and indeed most, published/reported work in the area of metabolic engineering has focused on the production of single or small groups of related, generally known compounds through manipulation of the corresponding specific biosynthetic pathway. However, this limits the utility of the developed system for investigation of novel metabolism. We wish to utilize metabolic engineering instead for biosynthetic pathway elucidation and enzymatic investigation via facile incorporation of putative biosynthetic enzymes and characterization of the resulting novel metabolites.
Our interest centers on the large superfamily of labdane-related diterpenoids, which is comprised of almost 7,000 known natural products. Many of these are exclusively found in plants, whose large and complex genomes preclude ready identification of the corresponding enzymatic genes by physical proximity or on the basis of homology (particularly in the case of multigenic families). To provide a platform for facile investigation of novel enzymatic activity, we have developed a modular system for metabolically engineering E. coli to produce labdane-related diterpenes (Cyr et al. 2007). This provides an elegant approach to elucidation of diterpenoid biosynthetic pathways and corresponding enzymatic investigation and one that is amendable, with slight adaptation, to the study of any type of terpenoid metabolism. However, in order to realize this potential, it must be possible to readily produce the multimilligram quantities of resulting metabolite(s) needed for structural analysis (i.e., by NMR).
Initial work on this system demonstrated the ability of E. coli to produce desired diterpenes, albeit in small quantities of <100 µg/l of culture, presumably due to the limited endogenous isoprenoid precursor pools (Cyr et al. 2007). Here, we have set out to shift carbon flux from E. coli central metabolism for milligram production of diterpenes in liter scale culture growths. Two basic approaches were investigated, either supplementation of the endogenous MEP isoprenoid precursor pathway via overexpression of key enzymatic genes or addition of a heterologous MEV isoprenoid precursor pathway (this latter either in whole or, with supplementation, in part). The ability of these alternative approaches to improve diterpene production was assessed by analysis of their impact on abietadiene yield from cells coexpressing GGPP and abietadiene synthases.
Engineering MEP pathway enzymatic gene overexpression was found to increase abietadiene production. Interestingly, while only idi improved yield on its own, further improvement was evident upon overexpression of idi with both dxs and dxr, although these actually decreased yield on their own and when paired with each other or idi (Table 1). As culture growth was unimpaired by any combination of MEP enzymes and only abietadiene diterpene product yield was affected, this suggests flux through the pathway may be most responsible for the varied outcome arising from different gene combinations. Further, these results suggest that the investigated MEP pathway enzymatic genes exert distributed metabolic control, specifically dxs and dxr. While the isomerization of DMAPP and IPP mediated by idi seems to be the initial rate-limiting step, further increases are dependent on overexpression of both dxs and dxr. Notably, the production of 1-deoxy-D-xylulose-5-phosphate (DXP) mediated by dxs is not unique to isoprenoid precursor production, which is initiated by the ensuing production of 2-C-methyl-D-erythitol-4-phosphate mediated by dxr. Thus, our results suggest that increased flux to DXP (i.e., from dxs overexpression) is shunted to alternative metabolic fates in the absence of increased dxr activity, although overexpression of dxr alone also is not sufficient on its own to compete with the alternative pathways.
Introduction of a heterologous MEV pathway, either in part (with the appropriate supplementation) or whole, increased abietadiene yield beyond that observed with engineering of the endogenous MEP pathway (Table 1). These increases were particularly significant in bioreactor grown cultures, where yields >100 mg/L were observed, representing a >1,000-fold increase over that previously reported. However, this requires either costly mevalonate supplementation or the use of two plasmids for introduction of the whole MEP pathway, which limits the number of downstream steps that can be incorporated. In addition, obtaining truly significant increases further required use of a bioreactor. Nevertheless, baffled shake flask growth conditions with even the single plasmid associated with optimal MEP pathway engineering produces >10 mg/L of diterpene, providing a simple and economical means for obtaining the multimilligram quantities of novel metabolites necessary for their structural analysis.
Critically, the improvements reported here have been directly applied to the further incorporation of additional enzymatic genes (i.e., pairs of diterpene synthases rather than the bifunctional AgAS), carried on yet another plasmid, also with yields exceeding 100 mg/L in bioreactor fermentations. Thus, it should be possible to use the developed modular metabolic engineering system, with the yield improvements reported here, to investigate multistep biosynthetic pathways. We reiterate here that these include not only the labdane-related diterpenes that are the focus of our research, but other diterpenoid natural products and with slight modification (i.e., replacement of the GGPP synthase), other types of terpenoids as well.
Materials and methods
General
All molecular biology reactions were performed according to standard protocols. Polymerase chain reaction (PCR) was performed using Accuprime Pfx (Invitrogen), ligations were performed using T4 ligase (New England Biolabs), restriction endonuclease cleavage by New England Biolabs protocols, DNA preparations and gel extractions by Quiagen kits, and LR recombinations and entry cloning was performed by standard Gateway technology protocols (Invitrogen). Plasmid pGGAS was a pACYCDuet vector housing the GGPP synthase and abietadiene synthase from A. grandis in the first and second multiple cloning sites (MCS), respectively, as previously described (Cyr et al. 2007). All vectors and genes were verified by sequencing.
Design of vectors
The DUET family of vectors (Novagen) contain two MCS, each with its own T7 promoter and ribosome-binding site optimally positioned before the inserts. Incorporation of DESTination cassettes (Invitrogen) allows essentially any plasmid to be converted into a ligation-free module vector to allow for facile transfer of any gene into expression plasmids using the simple Gateway LR technology (Invitrogen). For our modular approach, a DESTination cassette containing attR1 recombination site, ccdB lethality selection gene, chloramphenical resistance gene, and attR2 recombination site, which allows for easier ligase-free att site recombination with removal of cassette selection genes, was inserted just downstream of the T7 promoter in the first multiple cloning site (MCS 1) in DUET vectors (Table 2). Construction of DEST cassette containing vectors was carried out via standard PCR, restriction enzyme, and ligation protocols, using NcoI and NotI sites. This DEST cassette allows quick transfer of any gene from a pENTR-containing entry vector (Gateway) into the DESTination-converted expression vector. As the use of DEST cassettes necessitates a long sequence (>50 bp) prior to the start codon, we utilize pENTR/SD entry clones, which have a Shine–Delgarno site optimally positioned before the start codon. All DEST containing Duet vectors were made in the same fashion (Table 3).
Vectors used for the MEV pathway were a kind gift from Prof. Jay Keasling (Martin et al. 2003). In their pMevT, the first three genes of the yeast MEV pathway, atoB, hmgs, and hmgr, were assembled into a synthetic operon. The latter half of the MEV pathway consists of the remaining three genes and IPP isomerase, which were also assembled into a synthetic operon to construct pMBI.
Plasmid construction
Construction of the MEP pathway combinations was done in pCDFDuet (Novagen), and all genes were cloned from E. coli. All genes contained in frame native stop codons to eliminate any C-terminal fusion tags from vectors. In all cases, NcoI and NotI were used for cloning into the MCS 1 site, whereas NdeI and XhoI were used for cloning into the MCS 2 site. These sites eliminate any N-terminal fusion tags from the vectors. This was performed using standard PCR protocols, restriction enzyme digest, and T4 ligation. All constructs were verified by complete sequencing of the insert, with inclusion of the relevant ribosome-binding sites. Plasmid pIDI consisted of the E. coli idi gene inserted into MCS 2. Plasmid pDXS consisted of dxs inserted into MCS 1. Plasmid pDXR consisted of dxr inserted into MCS 2. Plasmid pDR consisted of dxs inserted into MCS 1 and dxr inserted into MCS 2. Plasmid pIS consisted of idi inserted into MCS 2 and dxs inserted into MCS 1. Plasmid pIR consisted of idi inserted into MCS 1 and dxr inserted into MCS 2. Plasmid pIRS consisted of an MCS 2 two-gene operon of idi and dxr with a ribosome-binding site and eight nucleotides in-between, while dxs was inserted into MCS 1.
Construction of the pMevT/tHMGR duet vector, here called pMTH, was performed using a pETDuet base vector (Novagen) with a truncated copy of yeast HMGR ligated into MCS 2 using NdeI and KpnI sites, following incorporation of cut sites by PCR onto tHMGR using pMEVT as template. Within MCS 1 of pETDuet, a DEST cassette (Invitrogen) was ligated as previously noted, thus creating pETDuetDEST. PCR of the entire MevT operon using standard protocols was used for TOPO cloning to create a pENTR plasmid (Invitrogen) housing the entire operon, which was verified by complete sequencing. The MevT operon was inserted into the DEST cassette of the pCDFDuetDEST vector using standard LR recombination.
The pMBID plasmid was constructed in a similar fashion as pMBIS, as described by Martin et al. (2003). Briefly, SacI and SacII sites were incorporated by PCR at the 5′ and 3′ ends, respectively, of a GGPP synthase from A. grandis (Burke and Croteau 2002), enabling its insertion into the MBI operon, which was verified by complete sequencing.
Culture growth
All media recipes were made at pH 7 and autoclaved with the appropriate starting ingredients. NZY media for 1 L was made with 5 g yeast extract, 10 g casein hydrolysate, 5 g NaCL, and 1 g MgSO4. NZYB consisted of NZY plus phosphate buffer. TB media for 1 L was made containing 12 g yeast extract, 24 g casein hydrolysate, 4 mL glycerol, and phosphate buffer. FL media for 1 L was comprised of 10 g yeast extract, 20 g casein hydrolysate, 5 g NaCL, 1 g MgSO4, 20 mL glycerol, and phosphate buffer. Supplementation with glycerol and phosphate buffer occurred at the time of induction.
All transformations were carried out using E. coli strain C41 Overexpress (Lucigen), an optimal strain for terpene synthase expression (Prisic and Peters 2007). Transformations consisting of three or more plasmids required 0.5 µg plasmid DNA in 50 µL aliquots of chemically competent E. coli C41 cells. The resulting recombinant strains were grown under selective conditions using the appropriate antibiotics at concentrations of 25 µg/mL for carbenicillin, 20 µg/mL for chloramphenicol, 15 µg/mL tetracycline, and 15 µg/mL spectinomycin. For all concentrations listed, negative controls lacking resistance to one marker confirmed the ability of the lower antibiotic concentrations to fully inhibit growth. Expression growths were initiated from several colonies inoculated together into liquid media. For all shake flasks cultures, initial growth to log phase was carried out at 37 °C (A600 ∼0.6), with the temperature then dropped to 16 °C, whereupon the pH was adjusted to 7.0 and phosphate buffer and glycerol, where described, were added. Shaking was held at 200 rpm, and the cultures induced 1 h after dropping the temperature to 16 °C by the addition of IPTG to 1 mM. Baffled flasks were tested under the same conditions as regular Erlenmeyer flasks.
During growth, cultures were intermittently monitored for pH, cell density, and where applicable, product output (as indicated in the various figures). Cell density was monitored by absorbance of aliquots (one in ten dilutions) at 600 nm in a Varian spectrophotometer. Culture pH was monitored throughout growth and adjusted, if necessary, to ensure that the culture pH remained in the 6.5 to 7.5 range, although the cultures rarely required adjustments after the shift to 16 °C. Supplementation with pyruvate or mevalonolactone was administered via pulse feeding with 1 M solutions at regular intervals of 12 h for the first 36 h, up to the desired maximum concentration. Investigation of the role of cell density on product formation (Fig. 3) was conducted in 48 h parallel aliquots for each density, from the time of IPTG induction, whereby the cell densities were controlled by moving the culture in aliquots from the initial incubation temperature of 37 to 16 °C, where it was held for the remainder of the time course. The values recorded for the cell density are the average of the two aliquots; the percent variance between the duplicate cell densities was smaller than the percent variance between the determined abietadiene yield, therefore error in abietadiene yield is displayed. Note that the data reported in Tables 1 and 2, as well as Figs. 2, 3, 4, 5, 7, and 8, were derived from parallel fermentation runs (i.e., all the data reported in each of these came from cultures grown together in the same incubator, each in duplicate).
Bioreactor growths were conducted in a New Brunswick BioFlo110 fermentor, set up according the manufacturer's instructions. Under our settings, the cultures were stirred at 300 rpm, their temperature maintained at 20 °C, pH held at 7.2 (using 5 M KOH and 5 M HCL reservoirs connected to the A and B pumps), chemically resistant tubing was used, and air flow was maintained at 4 lpm using triple micron filtered air. Pulse feeding supplements were introduced through the feeding septa, and cell density and product output monitored, as well as pH verified, by intermittent sample removal for subsequent measurement.
Product analysis
Abietadiene analysis consisted of hexane extraction, followed by verification and quantification by gas chromatography (GC) with flame ionization detection (FID) or mass spectrometry (MS) analyses, as described previously (Cyr et al. 2007). Throughout the extractions, glass was exclusively used, as plastic may result in absorption of hydrophobic diterpenes. Preliminary work demonstrated that extraction of entire cultures (media and cells together) resulted in the same level of extraction efficiency as cultures that were separated by centrifugation in glass tubes into cell pellet and media followed by separate extraction of the separated cell pellet and media. Mixed phase cultures or extractions utilizing hydrophobic beads (Sepabeads SP850, Supelco) did not result in an increase in product extraction or production. Additionally, hexane was shown to be a suitable organic solvent for extraction as other solvents, such as ethyl acetate, or mixtures of solvents resulted in the same efficiency of extraction as hexane, which has the advantage of being more easily evaporated under nitrogen gas.
At the completion of growth, cultures were lysed by brief sonication, and hexane was added in an equal volume (i.e., for 50 mL cultures, 50 mL of hexane was overlayed). The lysed cultures were then vigorously shaken multiple times for thorough extraction and then allowed to settle overnight at 4 °C. Additionally, sonication was verified to not cause diterpene rearrangement or degradation. To assist in emulsion removal in the organic phase, 2% (volumer per volume) amount of ethanol was added to disrupt surface tension of the emulsion and clear the organic phase. In all cases, emulsions were agitated until they comprised less than 20% of the total mixture volume; addition of acid will reduce emulsions even further, till they comprise a minimal volume as evidenced by a thin layer between phases. Simple extraction with hexane routinely resulted in diterpene isolation that was >90% pure as determined by GC-FID. In cases where alcohol derivatives emerged, presumably caused by dephosphorylation of prenyl diphosphate intermediates, as identified by GC-MS, passage over silica gel columns absorbed the oxygen containing byproducts while permitting the diterpenes to pass through, although the mixture of abietadiene double bond isomers also then undergoes rearrangement to the most stable abieta-7,13-diene form (Peters et al. 2000). A known volume aliquot of the organic overlay (10 mL in the case of 50 mL cultures) was removed and evaporated in glass test tubes under a stream of inert diatomic nitrogen gas. The sample was then resuspended in 1 mL of hexane for GC analysis.
Verification of abietadiene was provided by GC-FID and GC-MS comparison with authentic abietadiene produced by assays with purified, recombinant AgAS reacting with GGPP, as previously described (Wilderman and Peters 2007). Quantification was made by GC-FID peak area comparison to cembrene, a commercially available diterpene hydrocarbon, as well as kaurene, a synthetic standard of known quantity; each demonstrated identical linear agreement of peak area in quantities ranging from 5 to 1000 ng for cembrene and 5 to 100 ng for kaurene. Additionally, it was later found that sonication of cultures did not improve extraction efficiency, with extraction of unlysed (i.e., nonsonicated) culture offering the added benefit of decreased emulsion formation. However, to maintain consistency throughout, all measurements reported in this manuscript were from cultures that had been sonicated prior to extraction.
References
Burke C, Croteau R (2002) Interaction with the small subunit of geranyl diphosphate synthase modifies the chain length specificity of geranylgeranyl diphosphate synthase to produce geranyl diphosphate. J Biol Chem 277(5):3141–3149
Chang MCY, Eachus RA, Trieu W, Ro D-K, Keasling JD (2007) Engineering Escherichia coli for production of functionalized terpenoids using plant P450s. Nat Chem Biol 3:274–277
Clardy J, Walsh C (2004) Lessons from natural molecules. Nature 432:829–837
Cyr A, Wilderman PR, Determan M, Peters RJ (2007) A modular approach for facile biosynthesis of labdane-related diterpenes. J Am Chem Soc 129:6684–6685
Das A, Yoon SH, Lee SH, Kim JY, Oh DK, Kim SW (2007) An update on microbial carotenoid production: application of recent metabolic engineering tools. Appl Microbiol Biotechnol 77:505–512
Firn RD, Jones CG (2003) Natural products—a simple model to explain chemical diversity. Nat Prod Rep 20:382–391
Harris LJ, Saparno A, Johnston A, Prisic S, Xu M, Allard S, Kathiresan A, Ouellet T, Peters RJ (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent-copalyl diphosphate synthase. Plant Mol Biol 59:881–894
Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD (2003) Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21(7):796–802
McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7:1015–1026
Morrone D, Xu M, Fulton DB, Determan MK, Peters RJ (2008) Increasing complexity of a diterpene synthase reaction with a single residue switch. J Am Chem Soc 130:5400–5401
Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ (2009) Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583(2):475–480
Newman JD, Cragg GM (2007) Natural products as sources of new drugs over the last 25 years. J Nat Prod 70:461–477
Newman JD, Marshall J, Chang M, Nowroozi R, Paradise E, Pitera D, Newman KL, Keasling JD (2006) High-level production of amorpha-4, 11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol Bioeng 95(4):684–691
Peters RJ, Flory JE, Jetter R, Ravn MM, Lee H-J, Coates RM, Croteau RB (2000) Abietadiene synthase from grand fir (Abies grandis): characterization and mechanism of action of the "pseudomature" recombinant enzyme. Biochemistry 39(50):15592–15602
Pitera D, Paddon CJ, Newman JD, Keasling JD (2007) Balancing a heterologous mevalonate pathway for improved isoprenoid production in Escherichia coli. Metab Eng 9:193–207
Prisic S, Peters RJ (2007) Synergistic substrate inhibition of ent-copalyl diphosphate synthase: a potential feed-forward inhibition mechanism limiting gibberellin metabolism. Plant Physiol 144:445–454
Prisic S, Xu M, Wilderman PR, Peters RJ (2004) Rice contains disparate ent-copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol 136(4):4228–4236
Rohmer M (2008) From molecular fossils of bacterial hopanoids to the formation of isoprene units: discovery and elucidation of the methylerythritol phosphate pathway. Lipids 43:1095–1107
Wilderman PR, Peters RJ (2007) A single residue switch converts abietadiene synthase into a pimaradiene-specific cyclase. J Am Chem Soc 129:15736–15737
Xu M, Hillwig ML, Prisic S, Coates RM, Peters RJ (2004) Functional identification of rice syn-copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J 39(3):309–318
Acknowledgments
We thank Prof. Jay Keasling (UC Berkeley) for the kind gift of pMevT and pMBI. This work was supported by a grant from the NIH (GM086281) to R.J.P., and M.K.D. was supported, in part, by an REU supplement to NSF grant MCB-0416948 held by R.J.P.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Information
(PDF 875 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Morrone, D., Lowry, L., Determan, M.K. et al. Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl Microbiol Biotechnol 85, 1893–1906 (2010). https://doi.org/10.1007/s00253-009-2219-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2219-x