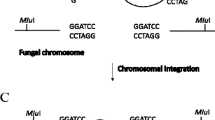
Abstract
Filamentous fungi have a long-standing tradition as industrial producers of primary and secondary metabolites. Initially, industrial scientists selected production strains from natural isolates that fulfilled both microbiological and technical requirements for economical production processes. Subsequently, genetically modified strains with novel properties were obtained through traditional strain improvement programs relying mostly on random mutagenesis. In recent years, however, recombinant technologies have contributed significantly to improve the capacities of production and have also allowed the design of genetically manipulated strains. These major advances were only made possible by basic research bringing deeper and novel insights into cellular and molecular fungal processes, thus allowing the design of genetically manipulated strains. This better understanding of fundamental genetic processes in model organisms has resulted in the design and generation of new experimental transformation strategies to manipulate specifically gene expression and function in diverse filamentous fungi, including those having a biotechnical significance. In this review, we summarize recent developments in the application of homologous DNA recombination and RNA interference to manipulate fungal recipients for further improvement of physiology and development in regards to biotechnical and pharmaceutical applications.




Similar content being viewed by others
References
Barton LM, Prade RA (2008) Inducible RNA interference of brlAbeta in Aspergillus nidulans. Eukaryot Cell 7:2004–2007
Cardoza RE, Vizcaíno JA, Hermosa MR, Sousa S, González FJ, Llobell A, Monte E, Gutiérrez S (2006) Cloning and characterization of the erg1 gene of Trichoderma harzianum: effect of the erg1 silencing on ergosterol biosynthesis and resistance to terbinafine. Fungal Genet Biol 43:164–178
Casqueiro J, Gutierrez S, Banuelos O, Hijarrubia MJ, Martin JF (1999) Gene targeting in Penicillium chrysogenum: disruption of the lys2 gene leads to penicillin overproduction. J Bacteriol 181:1181–1188
Catlett NL, Lee BN, Yoder OC, Turgeon BG (2003) Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet Newsl 50:9–11
Choquer M, Robin G, Le Pecheur P, Giraud C, Levis C, Viaud M (2008) Ku70 or Ku80 deficiencies in the fungus Botrytis cinerea facilitate targeting of genes that are hard to knock out in a wild-type context. FEMS Microbiol Lett 289:225–232
Cogoni C, Macino G (1999a) Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286:2342–2344
Cogoni C, Macino G (1999b) Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399:166–169
Cogoni C, Irelan JT, Schumacher M, Schmidhauser TJ, Selker EU, Macino G (1996) Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J 15:3153–3163
Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci USA 103:10352–10357
Costa AM, Mills PR, Bailey AM, Foster GD, Challen MP (2008) Oligonucleotide sequences forming short self-complimentary hairpins can expedite the down-regulation of Coprinopsis cinerea genes. J Microbiol Methods 75:205–208
Critchlow SE, Jackson SP (1998) DNA end-joining: from yeast to man. Trends Biochem Sci 23:394–398
de Backer MD, Raponi M, Arndt GM (2002) RNA-mediated gene silencing in non- pathogenic and pathogenic fungi. Curr Opin Microbiol 5:323–329
da Silva Ferreira ME, Kress MR, Savoldi M, Goldman MH, Hartl A, Heinekamp T, Brakhage AA, Goldman GH (2006) The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:207–211
Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39:431–451
de Hoogt R, Luyten WHML, Contreras R, De Backer MD (2000) PCR- and ligation-mediated synthesis of split-marker cassettes with long flanking homology regions for gene disruption in Candida albicans. Biotechniques 28:1112–1116
de Jong JF, Deelstra HJ, Wosten HA, Lugones LG (2006) RNA-mediated gene silencing in monokaryons and dikaryons of Schizophyllum commune. Appl Environ Microbiol 72:1267–1269
El-Khoury R, Sellem CH, Coppin E, Boivin A, Maas MFPM, Debuchy R, Sainsard-Chanet A (2008) Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr Genet 53:249–258
Fairhead C, Llorente B, Denis F, Soler M, Dujon B (1996) New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using 'split-marker' recombination. Yeast 12:1439–1457
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Fitzgerald A, Van Kan JA, Plummer KM (2004) Simultaneous silencing of multiple genes in the apple scab fungus, Venturia inaequalis, by expression of RNA with chimeric inverted repeats. Fungal Genet Biol 41:963–971
Fu J, Hettler E, Wickes BL (2006) Split marker transformation increases homologous integration frequency in Cryptococcus neoformans. Fungal Genet Biol 43:200–212
Goins CL, Gerik KJ, Lodge JK (2006) Improvements to gene deletion in the fungal pathogen Cryptococcus neoformans: Absence of Ku proteins increases homologous recombination, and co-transformation of independent DNA molecules allows rapid complementation of deletion phenotypes. Fungal Genet Biol 43:531–544
Goldoni M, Azzalin G, Macino G, Cogoni C (2004) Efficient gene silencing by expression of double stranded RNA in Neurospora crassa. Fungal Genet Biol 41:1016–1024
Gross H (2009) Genomic mining-a concept for the discovery of new bioactive natural products. Curr Opin Drug Discov Devel 12:207–219
Guangtao Z, Hartl L, Schuster A, Polak S, Schmoll M, Wang TH, Seidl V, Seiboth B (2009) Gene targeting in a nonhomologous end joining deficient Hypocrea jecorina. J Biotechnol 139:146–151
Haarmann T, Lorenz N, Tudzynski P (2008) Use of a nonhomologous end joining deficient strain (∆ku70) of the ergot fungus Claviceps purpurea for identification of a nonribosomal peptide synthetase gene involved in ergotamine biosynthesis. Fungal Genet Biol 45:35–44
Hammond TM, Keller NP (2005) RNA silencing in Aspergillus nidulans is independent of RNA-dependent RNA polymerases. Genetics 169:607–617
Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 4:639–648
Heneghan MN, Costa AMSB, Challen MP, Mills PR, Bailey A, Foster GD (2007) A comparison of methods for successful triggering of gene silencing in Coprinus cinereus. Mol Biotechnol 35:283–294
Henry C, Mouyna I, Latge JP (2007) Testing the efficacy of RNA interference constructs in Aspergillus fumigatus. Curr Genet 51:277–284
Hoff B, Kamerewerd J, Sigl C, Zadra I, Kück U (2009) Homologous recombination in the antibiotic producer Penicillium chrysogenum: strain ∆Pcku70 shows up-regulation of genes from the HOG pathway. Appl Microbiol Biotechnol 85:1081–1094
Hsu HL, Yannone SM, Chen DJ (2002) Defining interactions between DNA-PK and ligase IV/XRCC4. DNA Repair (Amst) 1:225–235
Ishibashi K, Suzuki K, Ando Y, Takakura C, Inoue H (2006) Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc Natl Acad Sci USA 103:14871–14876
Janus D, Hoff B, Hofmann E, Kück U (2007) An efficient fungal RNA-silencing system using the DsRed reporter gene. Appl Environ Microbiol 73:962–970
Janus D, Hoff B, Kück U (2009) Evidence for Dicer-dependent RNA interference in the industrial penicillin producer Penicillium chrysogenum. Microbiology 155:3946–3956
Jeong JS, Mitchell TK, Dean RA (2007) The Magnaporthe grisea snodprot1 homolog, MSP1, is required for virulence. FEMS Microbiol Lett 273:157–165
Jin FJ, Takahashi T, Machida M, Koyama Y (2009) Identification of a bHLH-type transcription regulator gene by systematically deleting large chromosomal segments in Aspergillus oryzae. Appl Environ Microbiol 75:5943–5951
Kadotani N, Nakayashiki H, Tosa Y, Mayama S (2003) RNA silencing in the phytopathogenic fungus Magnaporthe oryzae. Mol Plant Microbe Interact 16:769–776
Khatri M, Rajam MV (2007) Targeting polyamines of Aspergillus nidulans by siRNA specific to fungal ornithine decarboxylase gene. Med Mycol 45:211–220
Kitamoto N, Yoshino S, Ohmiya K, Tsukagoshi N (1999) Sequence analysis, overexpression, and antisense inhibition of a beta-xylosidase gene, xylA, from Aspergillus oryzae KBN616. Appl Environ Microbiol 65:20–24
Kito H, Fujikawa T, Moriwaki A, Tomono A, Izawa M, Kamakura T, Ohashi M, Sato H, Abe K, Nishimura M (2008) MgLig4, a homolog of Neurospora crassa Mus-53 (DNA ligase IV), is involved in, but not essential for, non-homologous end-joining events in Magnaporthe grisea. Fungal Genet Biol 45:1543–1551
Krajaejun T, Gauthier GM, Rappleye CA, Sullivan TD, Klein BS (2007) Development and application of a green fluorescent protein sentinel system for identification of RNA interference in Blastomyces dermatitidis illuminates the role of septin in morphogenesis and sporulation. Eukaryot Cell 6:1299–1309
Krappmann S (2007) Gene targeting in filamentous fungi: the benefits of impaired repair. Fungal Biol Rev 21:25–29
Krappmann S, Sasse C, Braus GH (2006) Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot Cell 5:212–215
Larrondo LF, Colot HV, Baker CL, Loros JJ, Dunlap JC (2009) Fungal functional genomics: tunable knockout-knock-in expression and tagging strategies. Eukaryot Cell 8:800–804
Lee DW, Seong KY, Pratt RJ, Baker K, Aramayo R (2004) Properties of unpaired DNA required for efficient silencing in Neurospora crassa. Genetics 167:131–150
Liu G, Casqueiro J, Banuelos O, Cardoza RE, Gutierrez S, Martin JF (2001) Targeted inactivation of the mecB gene, encoding cystathionine-γ-lyase, shows that the reverse transsulfuration pathway is required for high-level cephalosporin biosynthesis in Acremonium chrysogenum C10 but not for methionine induction of the cephalosporin genes. J Bacteriol 183:1765–1772
Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL (2002) RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics 160:463–470
Martens H, Novotny J, Oberstrass J, Steck TL, Postlethwait P, Nellen W (2002) RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol Biol Cell 13:445–453
Maruyama J, Kitamoto K (2008) Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (∆ligD) in Aspergillus oryzae. Biotechnol Lett 30:1811–1817
Matityahu A, Hadar Y, Dosoretz CG, Belinky PA (2008) Gene silencing by RNA interference in the white rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol 74:5359–5365
McDonald T, Brown D, Keller NP, Hammond TM (2005) RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol Plant Microbe Interact 18:539–545
Meek K, Gupta S, Ramsden DA, Lees-Miller SP (2004) The DNA-dependent protein kinase: the director at the end. Immunol Rev 200:132–141
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of ligD (human lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol 45:878–889
Moriwaki A, Ueno M, Arase S, Kihara J (2007) RNA-mediated gene silencing in the phytopathogenic fungus Bipolaris oryzae. FEMS Microbiol Lett 269:85–89
Mouyna I, Henry C, Doering TL, Latge JP (2004) Gene silencing with RNA interference in the human pathogenic fungus Aspergillus fumigatus. FEMS Microbiol Lett 237:317–324
Nakayashiki H, Nguyen QB (2008) RNA interference: roles in fungal biology. Curr Opin Microbiol 11:494–502
Nakayashiki H, Kadotani N, Mayama S (2006) Evolution and diversification of RNA silencing proteins in fungi. J Mol Evol 63:127–135
Namekawa SH, Iwabata K, Sugawara H, Hamada FN, Koshiyama A, Chiku H, Kamada T, Sakaguchi K (2005) Knockdown of LIM15/DMC1 in the mushroom Coprinus cinereus by double-stranded RNA-mediated gene silencing. Microbiology 151:3669–3678
Naranjo L, Martin de Valmaseda E, Casqueiro J, Ullán RV, Lamas-Maceiras M, Banuelos O, Martin JF (2004) Inactivation of the lys7 gene, encoding saccharopine reductase in Penicillium chrysogenum, leads to accumulation of the secondary metabolite precursors piperideine-6-carboxylic acid and pipecolic acid from alpha-aminoadipic acid. Appl Environ Microbiol 70:1031–1039
Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR (2006) A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172:1557–1566
Ngiam C, Jeenes DJ, Punt PJ, Van Den Hondel CA, Archer DB (2000) Characterization of a foldase, protein disulfide isomerase A, in the protein secretory pathway of Aspergillus niger. Appl Environ Microbiol 66:775–782
Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, Nakayashiki H (2008) Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol 68:1348–1365
Nicolás FE, Torres-Martínez S, Ruiz-Vázquez RM (2003) Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J 22:3983–3991
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101:12248–12253
Palmbos PL, Daley JM, Wilson TE (2005) Mutations of the Yku80 C terminus and Xrs2 FHA domain specifically block yeast nonhomologous end joining. Mol Cell Biol 25:10782–10790
Pöggeler S, Kück U (2006) Highly efficient generation of signal transduction knockout mutants using a fungal strain deficient in the mammalian ku70 ortholog. Gene 378:1–10
Rappleye CA, Engle JT, Goldman WE (2004) RNA interference in Histoplasma capsulatum demonstrates a role for alpha-(1, 3)-glucan in virulence. Mol Microbiol 53:153–165
Renckens S, De Greve H, Van Montagu M, Hernalsteens JP (1992) Petunia plants escape from negative selection against a transgene by silencing the foreign DNA via methylation. Mol Gen Genet 233:53–64
Romano N, Macino G (1992) Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol 6:3343–3353
Schümann J, Hertweck C (2007) Molecular basis of cytochalasan biosynthesis in fungi: gene cluster analysis and evidence for the involvement of a PKS-NRPS hybrid synthase by RNA silencing. J Am Chem Soc 129:9564–9565
Shafran H, Miyara I, Eshed R, Prusky D, Sherman A (2008) Development of new tools for studying gene function in fungi based on the Gateway system. Fungal Genet Biol 45:1147–1154
Shiu PK, Raju NB, Zickler D, Metzenberg RL (2001) Meiotic silencing by unpaired DNA. Cell 107:905–916
Singh S, Braus-Stromeyer SA, Timpner C, Tran VT, Lohaus G, Reusche M, Knufer J, Teichmann T, von Tiedemann A, Braus GH (2009) Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross-pathway control in the xylem. Appl Microbiol Biotechnol. doi:10.1007/s00253-009-2269-0
Snoek ISI, van der Krogt ZA, Touw H, Kerkman R, Pronk JT, Bovenberg RAL, van den Berg MA, Daran JM (2009) Construction of an hdfA Penicillium chrysogenum strain impaired in non-homologous end-joining and analysis of its potential for functional analysis studies. Fungal Genet Biol 46:418–426
Takahashi T, Masuda T, Koyama Y (2006a) Identification and analysis of Ku70 and Ku80 homologs in the koji molds Aspergillus sojae and Aspergillus oryzae. Biosci Biotech Biochem 70:135–143
Takahashi T, Masuda T, Koyama Y (2006b) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Gen Genom 275:460–470
Takahashi T, Jin FJ, Sunagawa M, Machida M, Koyama Y (2008) Generation of large chromosomal deletions in koji molds Aspergillus oryzae and Aspergillus sojae via a Loop-Out recombination. Appl Environ Microbiol 74:7684–7693
Takahashi T, Jin FJ, Koyama Y (2009) Nonhomologous end-joining deficiency allows large chromosomal deletions to be produced by replacement-type recombination in Aspergillus oryzae. Fungal Genet Biol 46:815–824
Takeno S, Sakuradani E, Tomi A, Inohara-Ochiai M, Kawashima H, Ashikari T, Shimizu S (2005) Improvement of the fatty acid composition of an oil-producing filamentous fungus, Mortierella alpina 1S-4, through RNA interference with delta12-desaturase gene expression. Appl Environ Microbiol 71:5124–5128
Tanguay P, Bozza S, Breuil C (2006) Assessing RNAi frequency and efficiency in Ophiostoma floccosum and O. piceae. Fungal Genet Biol 43:804–812
Ullán RV, Godio RP, Teijeira F, Vaca I, Garcia-Estrada C, Feltrer R, Kosalkova K, Martin JF (2008) RNA-silencing in Penicillium chrysogenum and Acremonium chrysogenum: validation studies using β-lactam genes expression. J Microbiol Methods 75:209–218
van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291–299
Venard C, Kulshrestha S, Sweigard J, Nuckles E, Vaillancourt L (2008) The role of a fadA ortholog in the growth and development of Colletotrichum graminicola in vitro and in planta. Fungal Genet Biol 45:973–983
Villalba F, Collemare J, Landraud P, Lambou K, Brozek V, Cirer B, Morin D, Bruel C, Beffa R, Lebrun MH (2008) Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet Biol 45:68–75
Whisson SC, Avrova AO, Van West P, Jones JT (2005) A method for double-stranded RNA-mediated transient gene silencing in Phytophthora infestans. Mol Plant Pathol 6:153–163
You BJ, Lee MH, Chung KR (2009) Gene-specific disruption in the filamentous fungus Cercospora nicotianae using a split-marker approach. Arch Microbiol 191:615–622
Zheng XF, Kobayashi Y, Takeuchi M (1998) Construction of a low-serine-type-carboxypeptidase-producing mutant of Aspergillus oryzae by the expression of antisense RNA and its use as a host for heterologous protein secretion. Appl Microbiol Biotechnol 49:39–44
Acknowledgments
We express our thanks to Gabriele Frenßen-Schenkel for the artwork; Kirsten Schanzmann, Marion Wolf, and Christina Liß for preparing the manuscript; and Stefanie Mertens for help in preparing the photos. The experimental work of the authors is funded by the Christian Doppler Society (Vienna), the Deutsche Forschungsgemeinschaft (Bonn), and Sandoz GmbH (Kundl).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kück, U., Hoff, B. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol 86, 51–62 (2010). https://doi.org/10.1007/s00253-009-2416-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-009-2416-7