Abstract
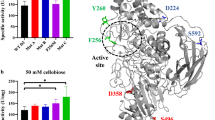
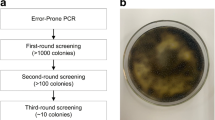
Cellulases can be engineered with enhanced properties for broad use in scientific and industrial applications. In this study, the wild-type cbh2 gene of the thermophilic fungus Chaetomium thermophilum encoding cellobiohydrolase II (CBHII) was mutagenized through in vitro directed evolution. The resulting Pichia pastoris yeast library was screened, and two transformants were selected for enhanced CBHII activities that were not attributed to increased gene copy numbers. The optimum fermentation times of the two mutant transformants were shortened to 4–5 days after methanol induction compared to 6 days for the wild-type. The optimum reaction temperature (60 °C) and pH level (5 or 6) of the mutant CBHII proteins, designated CBHIIX16 and CBHIIX305, were higher than those of wild-type CBHII (50 °C and pH 4). Kept at 80 °C for 1 h, CBHIIX16 and CBHIIX305 retained >50% of their activities, while the wild-type CBHII lost all activity. Sequence analysis of CBHIIX16 and CBHIIX305 revealed that they contained five and six mutated amino acids, respectively. Structural modeling confirmed the presence of carbohydrate binding type-1 and catalytic domains, where the hydrogen bond numbers between the 227th and 203rd amino acids were increased, which perhaps contributed to the elevated enzyme stability. Therefore, the two CBHII mutants selected for increased enzymatic activities also demonstrated elevated optimum reaction temperature and pH levels and enhanced thermal stability. These properties may be beneficial in practical applications for CBHII.





Similar content being viewed by others
References
Abuja PM, Pilz I, Claeyssens M, Tomme P (1988) Domain structure of cellobiohydrolase II as studied by small angle X-ray scattering: close resemblance to cellobiohydrolase I. Biochem Biophys Res Commun 156:180–185
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modeling. Bioinformatics 22:195–201
Arrizubieta MJ, Polaina J (2000) Increased thermal resistance and modification of the catalytic properties of a β-glucosidase by random mutagenesis and in vitro recombination. J Biol Chem 275:28843–28848
Bhat MK, Bhat S (1997) Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv 15:583–620
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72:248–254
Deshpande MV, Eriksson KE, Pettersson LG (1984) An assay for selective determination of exo-1,4-β-glucanase in a mixture of cellulolytic enzyme. Anal Biochem 138:481–487
Divne C, Ståhlberg J, Reinikainen T, Ruohonen L, Pettersson G, Knowles JK, Teeri TT, Jones TA (1994) The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science 265:524–528
Fan J, Gao X, Yang YW, Deng W, Li ZG (2007) Molecular cloning and characterization of a NAC-like gene in “Navel” orange fruit response to postharvest stresses. Plant Mol Biol Report 25:145–153
Ghose TK (1987) Measurement of cellulase activities. Pure Appl Chem 59:257–268
González-Blasco G, Sanz-Aparicio J, González B, Hermoso JA, Polaina J (2000) Directed evolution of β-glucosidase A from Paenibacillus polymyxa to thermal resistance. J Biol Chem 275:13708–13712
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Guo JP, Ma Y (2008) High-level expression, purification and characterization of recombinant Aspergillus oryzae alkaline protease in Pichia pastoris. Protein Expr Purif 58:301–308
Harvey AJ, Hrmova M, De Gori R, Varghese JN, Fincher GB (2000) Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 41:257–269
Heinzelman P, Snow CD, Wu I, Nguyen C, Villalobos A, Govindarajan S, Minshull J, Arnold FH (2009) A family of thermostable fungal cellulases created by structure-guided recombination. Proc Natl Acad Sci USA 106:5610–5615
Henrissat B, Claeyssens M, Tomme P, Lemesle L, Mornon JP (1989) Cellulase families revealed by hydrophobic cluster analysis. Gene 81:83–95
Hu P, Chase T Jr, Eveleigh DE (1993) Cloning of a Microbispora bispora cellobiohydrolase gene in Streptomyces lividans. Appl Microbiol Biotechnol 38:631–637
Iyayi CB, Bruchmann EE, Kubicek CP (1989) Induction of cellulase formation in Trichoderma reesei by cellobiono-1,5-lacton. Arch Microbiol 151:326–330
Kemppainen AJ, Shonnard DR (2005) Comparative life-cycle assessments for biomass-to-ethanol production from different regional feedstocks. Biotechnol Prog 21:1075–1084
Kim DW, Hong YG (2001) Description of cellobiohydrolases Ce16A and Ce17A from Trichoderma reesei using Langmuir-type models. Biotechnol Bioproc Eng 6:89–94
Kubicek CP (1981) Release of carboxymethyl-cellulase and β-glucosidase from cell walls of Trichoderma reesei. Eur J Appl Microbiol Biotechnol 13:226–231
Lantz SE, Goedegebuur F, Hommes R, Kaper T, Kelemen BR, Mitchinson C, Wallace L, Ståhlberg J, Larenas EA (2010) Hypocrea jecorina CEL6A protein engineering. Biotechnol Biofuels 3:20
Leisola M, Turunen O (2007) Protein engineering: opportunities and challenges. Appl Microbiol Biotechnol 75:1225–1232
Liang HK, Huang CM, Ko MT, Hwang JK (2005) Amino acid coupling patterns in thermophilic proteins. Proteins 59:58–63
Lima AO, Davis DF, Swiatek G, McCarthy JK, Yernool D, Pizzirani-Kleiner AA, Eveleigh DE (2009) Evaluation of GFP tag as a screening reporter in directed evolution of a hyperthermophilic β-glucosidase. Mol Biotechnol 42:205–215
Limam F, Chaabouni SE, Ghrir R, Marzouki N (1995) Two cellobiohydrolases of Penicillium occitanis mutant Pol 6: purification and properties. Enzyme Microb Technol 17:340–346
Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev 66:506–577
Mandels M (1985) Applications of cellulases. Biochem Soc Trans 13:414–416
Mattinen ML, Kontteli M, Kerovuo J, Linder M, Annila A, Lindeberg G, Reinikainen T, Drakenberg T (1997) Three-dimensional structures of three engineered cellulose-binding domains of cellobiohydrolase I from Trichoderma reesei. Protein Sci 6:294–303
McCarthy JK, Uzelac A, Davis DF, Eveleigh DE (2004) Improved catalytic efficiency and active site modification of 1,4-β-D-glucan glucohydrolase A from Thermotoga neapolitana by directed evolution. J Biol Chem 279:11495–11502
Medve J, Lee D, Tjerneld F (1998) Ion-exchange chromatographic purification and quantitative analysis of Trichoderma reesei cellulases cellobiohydrolase I, II and endoglucanase II by fast protein liquid chromatography. J Chromatogr A 808:153–165
Merril CR, Goldman D, Sedman SA, Ebert MH (1981) Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science 211:1437–1438
Moore JC, Jin HM, Kuchner O, Arnold FH (1997) Strategies for the in vitro evolution of protein function: enzyme evolution by random recombination of improved sequences. J Mol Biol 272:336–347
Murray PG, Collins CM, Grassick A, Tuohy MG (2003) Molecular cloning, transcriptional, and expression analysis of the first cellulase gene (cbh2), encoding cellobiohydrolase II, from the moderately thermophilic fungus Talaromyces emersonii and structure prediction of the gene product. Biochem Biophys Res Commun 301:280–286
Oksanen T, Pere J, Buchert J, Viikari L (1997) The effect of Trichoderma reesei cellulases and hemicellulases on the paper technical properties of never-dried bleached kraft pulp. Cellulose 4:329–339
Post CB, Ray WJ Jr (1995) Reexamination of induced fit as a determinant of substrate specificity in enzymatic reactions. Biochemistry 34:15881–15885
Rahman Z, Shida Y, Furukawa T, Suzuki Y, Okada H, Ogasawara W, Morikawa Y (2009) Evaluation and characterization of Trichoderma reesei cellulase and xylanase promoters. Appl Microbiol Biotechnol 82:899–908
Reinikainen T, Henriksson K, Siika-aho M, Teleman O, Poutanen K (1995) Low-level endoglucanase contamination in a Trichoderma reesei cellobiohydrolase II preparation affects its enzymatic activity on β-glucan. Enzyme Microb Technol 17:888–892
Rouvinen J, Bergfors T, Teeri T, Knowles JKC, Jones TA (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249:380–386
Rubingh DN (1997) Protein engineering from a bioindustrial point of view. Curr Opin Biotechnol 8:417–422
Sarkar A, Upadhyay SN (1993) Purification and characterization of cellulase from Bacillus thermoalcaliphilus isolated from a termite mound. Folia Microbiol 38:29–32
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Somogyi M (1952) Notes on sugar determination. J Biol Chem 195:19–23
Spezio M, Wilson DB, Karplus PA (1993) Crystal structure of the catalytic domain of a thermophilic endocellulase. Biochemistry 32:9906–9916
Sternberg D, Mandels GR (1979) Induction of cellulolytic enzymes in Trichoderma reesei by sophorose. J Bacteriol 139:761–769
Sun XY, Liu ZY, Zheng K, Song X, Qu YB (2008) The composition of basal and induced cellulase systems in Penicillium decumbens under induction or repression conditions. Enzyme Microb Technol 42:560–567
Urbanek H, Zalewska-Sobczak J, Borowifiska A (1978) Isolation and properties of extracellular cellulase–hemicellulase complex of Phoma hibernica. Arch Microbiol 118:265–269
Varrot A, Schülein M, Davies GJ (1999) Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding. Biochemistry 38:8884–8891
Wang GL, Gao PJ (1999) PCR-mediated analysis of transcription of CBH I and II genes from Trichoderma pseudokoningii and Penicillium janthinellum. Biotechnol Lett 21:321–324
Wang T, Liu X, Yu Q, Zhang X, Qu Y, Gao P, Wang T (2005) Directed evolution for engineering pH profile of endoglucanase III from Trichoderma reesei. Biomol Eng 22:89–94
Wohlfahrt G, Pellikka T, Boer H, Teeri TT, Koivula A (2003) Probing pH-dependent functional elements in proteins: modification of carboxylic acid pairs in Trichoderma reesei cellobiohydrolase Cel6A. Biochemistry 42:10095–10103
Wood TM, McCrae SI, Bhat KM (1989) The mechanism of fungal cellulase action. Synergism between enzyme components of Penicillium pinophilum cellulase in solubilizing hydrogen bond-ordered cellulose. Biochem J 260:37–43
Zeilinger S, Ebner A, Marosits T, Mach R, Kubicek CP (2001) The Hypocrea jecorina HAP 2/3/5 protein complex binds to the inverted CCAAT-box (ATTGG) within the cbh2 (cellobiohydrolase II-gene) activating element. Mol Genet Genomics 266:56–63
Acknowledgments
This work was funded by National High Technology Research and Development Program of China (863 Program) and National Natural Science Foundation of China (30571246).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, XJ., Peng, YJ., Zhang, LQ. et al. Directed evolution and structural prediction of cellobiohydrolase II from the thermophilic fungus Chaetomium thermophilum . Appl Microbiol Biotechnol 95, 1469–1478 (2012). https://doi.org/10.1007/s00253-011-3799-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-011-3799-9