Abstract
Flow cytometry (FCM) is widely used in cancer research for diagnosis, detection of minimal residual disease, as well as immune monitoring and profiling following immunotherapy. In all these applications, the challenge is to detect extremely rare cell subsets while avoiding spurious positive events. To achieve this objective, it helps to be able to analyze FCM data using multiple markers simultaneously, since the additional information provided often helps to minimize the number of false positive and false negative events, hence increasing both sensitivity and specificity. However, with manual gating, at most two markers can be examined in a single dot plot, and a sequential strategy is often used. As the sequential strategy discards events that fall outside preceding gates at each stage, the effectiveness of the strategy is difficult to evaluate without laborious and painstaking back-gating. Model-based analysis is a promising computational technique that works using information from all marker dimensions simultaneously, and offers an alternative approach to flow analysis that can usefully complement manual gating in the design of optimal gating strategies. Results from model-based analysis will be illustrated with examples from FCM assays commonly used in cancer immunotherapy laboratories.
Similar content being viewed by others
Flow cytometry in immune monitoring for cancer immunotherapy
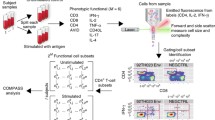
Flow cytometry (FCM) is one of the older high throughput techniques, and as such has become an integral tool in immune monitoring. In FCM, cells in a fluid medium are streamed in a single file through a capillary tube, and light from several lasers is directed at each individual cell as it passes through. Typically, each cell is labeled with different fluorescent dyes attached to monoclonal antibodies that recognize specific cellular markers that may be cell surface or intracellular proteins. Laser light hitting each cell results in scatter and activation of the fluorescent dyes bound to the cell. The light from forward and side scatter are detected electronically in separate channels, as is the light emitted by each fluorescent dye molecule as it relaxes from its activated state. Since each monoclonal antibody of a given specificity is bound to a different fluorescent dye, the amount of light in each wavelength detected will be directly proportional to the number of bound antibodies of that specificity, and therefore a measure of the density of the cellular marker targeted by that monoclonal antibody on each cell [1].
Flow cytometry assays therefore provide the ability to measure cellular marker levels for individual cells. With flow rates of hundreds to thousands of cells per second, FCM also captures cellular population statistics in a single assay. FCM can thus measure both the phenotype and function of specific cell subtypes in a sample that may contain many different cell subtypes, unlike assays like microarrays and proteomics that measure aggregate features of the entire mixture of cells. This ability of FCM to identify and characterize rare cell subsets is particularly critical in monitoring the immune response following cancer immunotherapy and detection of minimal residual disease (MRD), where the cells of interest typically constitute much less than 1% of the total immune cells in the peripheral blood. In this review, we will use the context of immune monitoring for cancer immunotherapy for illustration, but the methods discussed should be widely applicable. In particular, our examples will focus on the tracking of antigen-specific lymphocytes by T cell receptor binding to specific fluorescence labeled peptide-MHC tetramers, as well as intracellular staining (ICS) and proliferation assays for functional readouts.
Challenges to manual analysis
For multi-center clinical trials, the ability to obtain reproducible and consistent results from FCM across different institutions is obviously critical. Consequently, there is a long history, spanning at least two decades, of efforts to enable robust and accurate enumeration of lymphocyte subsets, and in particular, to achieve the optimal balance of recovery of the populations of interest with minimal contamination from other cell subsets or debris. These efforts include the use of backgating for measuring purity and recovery combined with checksums [2], use of CD45 in three-color and four-color assays [3, 4], use of a single platform technology for absolute counts [5, 6], panleukogating [7] and the use of pre- aliquoted lyophilized reagents [8].
Throughout these studies, it has been recognized that different gating methods across institutions are a known major source of assay variability and that standardized analytical strategies improve both intra- and inter-laboratory variability [9]. In particular, the study by Maecker et al. [8] highlighted the contribution of gating as a major source of variability in a multi-center study of multi-parameter ICS assays. With the advent of polychromatic FCM technology [10], issues of standardization and variability due to gating techniques become even more acute, due to the exponential increase in analytical complexity with each marker added [11].
In traditional gating-based analysis, expert-defined boundaries are used to identify flow event subsets of interest [11–14]. Because gating is performed manually based on visual comparison with controls, such boundaries are constrained to be in one or two dimensions only, and a sequence of gates must be used to analyze multi-dimensional data sets. Limitations of software based on the gating concept are:
-
1.
Gating is expert-driven and different operators often disagree on the ideal gating strategy for a given panel
-
2.
Even with a common gating strategy, gating is difficult to replicate across different instruments and institutions [7, 8, 15, 16]
-
3.
Gating is a one- to two-dimensional procedure and difficult to scale to multi-parameter data sets
-
4.
Gating per se provides no guidance on the subsequent interpretation of the clinical relevance for the cell subsets found.
Because of these issues, it is highly beneficial for centers performing immunotherapy research for cancer to participate in proficiency testing, so that feedback on their performance relative to other clinical laboratories can be assessed. In the field of cancer vaccines, leadership in administering these proficiency tests is provided by the Cancer Immunotherapy Consortium (CIC) ImmunoAssay Working Group (IAWG) and the Cancer Immunoguiding Program (CIP) of the Cancer Immunotherapy Association (CIMT). These groups offer proficiency testing for ELISPOT as well as ICS and tetramer-based FCM assays, and have achieved significant gains in assay harmonization and improvement across multiple laboratories in a short time period [17–20].
Classification via flow should be objective and reproducible. Even when using a common gating strategy, achieving consistent and reproducible results requires a significant investment of time and training of not only gate placement but also protocols for preparing samples, and participation in group standardization/proficiency testing efforts. Where standard protocols do not exist (e.g. regulatory T cells), individual laboratories have to design their own gating strategies to identify the target cell subsets. In any event, achieving replicable results when the events of interest are exceptionally rare demands careful attention to potential false negatives and false positives, and the design of strategies to minimize them as much as possible.
Model-based analysis
Model-based analysis is an exciting alternative and supplement to gating-based analysis. In model-based analysis, flow event subsets are clustered by a data-driven algorithm and used to construct a generative probability model. Data fitting can be naturally performed in the full dimensionality of the data set, and hence scales well to high-dimensional data sets. In addition, expert input is not necessary for the analysis, removing the subjectivity inherent in gating-based approaches. In particular, because the modeling approach uses information from all markers simultaneously rather than pair-wise as in manual gating, it can often identify false negative and false positive events that are not easily discriminated in any two-dimensional projection. Model-based analysis therefore provides an objective, reproducible and scalable approach that directly addresses many of the limitations of gating-based analysis, and is hence under active investigation by several research groups [21–24].
Our approach to model-based analysis is based on the use of statistical mixture models [22, 25]. Statistical mixture models are very widely used in scientific problems where objects represented in several or many dimensions are to be clustered or classified. One appeal of mixture models is the ability to represent essentially any observed data distribution to a high degree of accuracy [26, 27]. In particular, mixture models have recently been demonstrated to be effective at identifying lymphocyte cell subsets from FCM data [22–24].
There has recently been renewed interest in the use of statistical mixture models for cell subset identification [21–24], including work by our group. Boedingheimer and Ferbas use a fixed component size mixture of Gaussians and an expectation maximization (EM) algorithm to identify differences in circulating B cells between normal donors and systemic lupus erythematosus (SLE) patients [21]. Lo et al. [23] also use an EM algorithm and fixed component size mixtures of Gaussian or t distributions to identify CD3+CD8+ cells in a graft-versus-host disease (GvHD) data set. Pyne et al. [24] use a finite mixture of skew and heavy-tailed multivariate distributions fitted with an EM algorithm and validated the approach on lymphoblastic cell lines and regulatory T cells. This recent work has clearly confirmed the validity and usefulness of a statistical mixture modeling approach for cell subset identification with FCM data. On a technical front, we have recently implemented mixture model algorithms that are optimized for massively parallel yet highly affordable graphics processing units (GPU) for speed-ups of two orders of magnitude, enabling the analysis of even massive data sets [28].
To fit an arbitrary n-dimensional flow data set, we require a flexible yet tractable probability model. One popular approach is to use a large number of well-understood multivariate normal distributions to build a complex and multi-modal distribution (see Fig. 1). With a sufficient number of normal components, such mixtures can fit any distribution arbitrarily well. With non-parametric mixture models, the number of mixture components needed for fitting can be directly estimated from the data itself, hence providing an automatic and objective basis for model fitting.
For interpretation of the model, however, we typically want to be able to quantify distinct cell subsets of interest. Because cell subsets may be asymmetric, they typically correspond to more than one normal component. We therefore require a way to merge normal components into clusters in such a way that each cluster corresponds to a distinct cell subset. One simple way to achieve this is to identify cell subsets with modes (peaks), and cluster all components that share the same mode as a single cell subset. Alternatively, the model can be specified as a mixture of mixtures, where the concept of a cell subset as comprising one or more normal components is directly built into the model [22].
Examples from tetramer, ICS and CFSE assays
We illustrate with an example using such mixture models to identify events positive for cytokine expression in an ICS assay, and how this approach can reduce false negative and positive events resulting in greater sensitivity and specificity (Fig. 2). A similar approach can be used to identify tetramer positive events (Fig. 3) and the characteristics of dividing and non-dividing lymphocytes in a CFSE assay (Fig. 4). These examples clearly show that model-based analysis provides new relevant information that can be used to improve gating strategies. Since mixture models identify cell subsets using all markers simultaneously, they more easily overcome issues from non-specific binding, such as binding to Fc receptors on macrophages or monocytes. As cells that bind non-specifically fall out as a separate cluster with multi-dimensional analysis, they are easily identified and can be removed from analysis (Fig. 2). However, because these non-specific events often fall in regions of interest, to find and exclude this cell subset manually generally requires a significant amount of time spent back-gating, looking where events in a region fall on other projections, or inclusion of more markers in the panel, such as viability dyes or dump channels. In addition, model-based analysis can identify potential cells of interest that are easily missed with manual gating, such as the ICS-positive CD4+ and CD8+ events in Fig. 2. Because of their atypical FSC and SSC characteristics, possibly due to their state of activation, they would be excluded in a standard gating strategy. While non-specific binding to Fc receptors on cell types such as macrophages and monocytes cannot be excluded, we believe that these CD3+CD4+ and CD3+CD8+ events may well be “blasting” lymphocytes. The ability to automatically find such cells enables model-based analysis to have improved sensitivity and specificity compared to manual analysis [22].
Mixture models reveal potential false negatives and false positives in an ICS assay. The left set of panels show ICS-positive events (small arrows) that are CD3+CD4+ or CD3+CD8+ but have atypical FSC/SSC characteristics possibly due to their state of activation that will be false negatives with a standard gating strategy. The middle panel shows a diagonal streak (large arrow) from non-specific binding due to dead cells. As this is classified as a separate cluster, it is trivial to eliminate such false positives with model-based analysis, but is otherwise extremely challenging with manual analysis. Right set of panels shows the relative frequency of ICS positive CD4+ and CD8+ cells obtained with manual analysis and automated clustering
Identification of tetramer-binding CD4-CD8+ cells with model-based analysis. Data are from an unpublished immune monitoring study of a multi-peptide vaccination trial designed for HLA-A2 patients with biochemical relapse after prostatectomy. The figure shows the frequency of cells binding to HLA-A2 tetramers (relative to CD4-CD8+ cells) constructed using an epitope derived from the prostate specific membrane antigen PSMA 711-719 ALFDIESKV before (top panel) and after (bottom panel) four vaccinations in one patient. Cells in both panels were expanded in vitro in the presence of the specific peptide and interleukins before staining. Background events are gray, CD4-CD8+ events are yellow and events from CD4-CD8+ tetramer+ are brown. Clusters were defined against the mean (μ) and standard deviations (SD) of all events from the before vaccination panel as follows: clusters were designated as CD4− if the mean CD4 of the cluster was less than μ-SD, CD8+ if the mean CD8 of the cluster was greater than μ + SD, and tetramer+ if the mean tetramer of the cluster was greater than μ + 3 SD
Traditional small, tight lymphocyte gates will exclude most dividing lymphocytes in a CFSE assay. Viable (amine low) CD3+CD4+ and CD3+CD8+ lymphocyte clusters are plotted to illustrate that proliferating CFSE-low cells (red) have highly atypical scatter characteristics and are likely to be excluded using standard “lymphocyte” gating strategies that are effective for non-proliferating lymphocytes (orange)
The mapping of modal clusters to cell subsets of interest is done by examining the statistical properties of each cluster. In Fig. 3, model-based analysis was applied to identify tetramer positive clusters using ongoing, still unpublished immune monitoring data from a novel multi-peptide vaccine study [29]. Data samples pre-vaccination were used as the negative control, and CD4−CD8+ tetramer+ clusters were identified from digitally concatenated data from PBMC taken before and after vaccination and expanded in vitro. Based on information from the negative control, two other heuristic criteria were also applied—the corresponding tetramer cluster in the negative control should have very few events, and the sample tetramer cluster should be significantly over-represented, over three fold with respect to the negative control. Once target clusters are identified, dot plots of the clusters in distinct colors are evaluated on all pair-wise marker combinations of scatters and markers, and reviewed by an experienced flow cytometrist. In Fig. 4, a model was fit to a CFSE assay. When looking at the FSC and SSC of clusters believed to correspond to proliferating CD4+ and CD8+ T cells it can be seen that the FSC and SSC of many proliferating cells would not fall within a standard “tight” lymphocyte gate and may be missed by an untrained eye using traditional gate based analysis, suggesting that model-based analysis may be more robust in some cases, especially for novel panels or atypical cell subsets.
In addition to cell subset identification, model-based approaches can also be used to quantify the uncertainty of any statistical inference made, as well as extract additional useful information from the fitted probability model. For example, fitted probability models may provide information on how to set a positivity threshold. Having fitted a probability model to the negative control, we can simulate many random draws from the fitted model, and hence determine a positivity threshold at any significance level desired. We are currently developing information measures based on the fitted model that tell us how much a given marker contributes to the identification of a specified cell subset, providing useful information to optimize panel design and gating strategy.
The high-dimensional future of flow cytometry
The latest generation of flow cytometers can resolve approximately 20 different colors [10]. This means that up to 20 different markers can be used to characterize the cells in a sample, and high-throughput setups with 96- or 384-well plates will generate large numbers of data sets, each potentially comprising 1–2 million events in 10–20 dimensions. Further increases in the number of markers that can be used in a single blood sample are currently limited by spectral emission overlap between different fluorochromes that reduces sensitivity.
While state-of-the-art FCM data samples are already of moderately high dimensionality, further increasing FCM resolution will allow substantial scientific and medical advances, including:
-
1.
Better discrimination of immune cell types on the basis of multiple activation, maturation, regulation, proliferation, and functional marker expression, increasing the likelihood of discovering biomarkers or flow cytometric signatures that correlate well with disease or vaccination outcomes;
-
2.
Screening of more peptide-MHC multimers for antigen-specific T cell populations in order to identify potential epitope targets for immunotherapy with fewer blood samples; and
-
3.
Better reconstruction of the kinetics of intracellular signaling with antibodies targeting phosphorylation sites of more proteins within signaling cascades.
Two major recent technological advances offer the potential to greatly increase the resolution of FCM. The first replaces fluorochrome dyes with stable isotope metal atoms as reporter tags, hence removing the problem of emission overlap [30]. Unlike traditional FCM, marker densities are evaluated by single cell mass spectrometry, hence ruling out live cell sorting applications. However, the resolution of individual reporter tags is greatly increased with this technology due to the lack of spectral overlap, and it is estimated that 60–100 markers are feasible. While this inductively coupled plasma mass spectrometry (ICP-MS) technology is relatively new, commercially available cytometers (CyTOF) and reagents (MAXPAR) have been manufactured by DVS Sciences (http://www.dvssciences.com).
The other major advance exploits combinatorial encoding of reporter tags to vastly increase the resolution of different cell subsets with a fixed number of markers [31, 32]. The basic idea is to use k colors to identify a single marker—in principle, if we can decode the color combinations efficiently, we can resolve n choose k different markers with n total colors and a k color per marker encoding. This dramatically increases the resolution of FCM—for example, the theoretical maximum number of different markers in a single blood sample that can be resolved with n = 11 and k = 2 is 55, rising with n = 18 and k = 9 to 48,620!
We have shown that modeling approaches to FCM analysis can help reduce false positive and false negative events, and hence contribute to the robust identification of very rare cell subsets critical in MRD and immune monitoring applications in cancer research. It is critical that such tools are properly validated before large-scale use, and we are currently working with the Cancer Immunoguiding Program (CIP) to compare manual and clustering methods for identifying tetramer positive cells. We have also recently been funded to validate model-based approaches for ICS FCM in collaboration with the Cancer Immunotherapy Consortium (CIC).
In the highly multiplexed future of FCM, such tools for efficient high-dimensional multivariate analysis of FCM data will become increasingly more essential. While in the past model-based analysis was slow, the ever increasing power of computers, including the use of general purpose GPU computing technologies that we have developed for FCM [28], are removing this limitation. We anticipate that software based on model-based analysis has the very real possibility of eventually displacing gating-based analysis software, due to improvements in sensitivity and specificity as well as the scalability, objectivity and automation brought by model-based analysis. This is especially true for the analysis of the high-volume, high-dimensional and multiplexed data sets that are increasingly important for identifying immunotherapeutic targets and for discovering immune correlates of vaccine efficacy or disease outcome in cancer patients.
References
Herzenberg LA, Tung J et al (2006) Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol 7(7):681–685
Schenker EL, Hultin LE et al (1993) Evaluation of a dual-color flow cytometry immunophenotyping panel in a multicenter quality assurance program. Cytometry 14(3):307–317
Nicholson J, Kidd P et al (1996) Three-color supplement to the NIAID DAIDS guideline for flow cytometric immunophenotyping. Cytometry 26(3):227–230
Schnizlein-Bick CT, Mandy FF et al (2002) Use of CD45 gating in three and four-color flow cytometric immunophenotyping: guideline from the National Institute of Allergy and Infectious Diseases, Division of AIDS. Cytometry 50(2):46–52
Reimann KA, O’Gorman MR et al (2000) Multisite comparison of CD4 and CD8 T-lymphocyte counting by single- versus multiple-platform methodologies: evaluation of Beckman Coulter flow-count fluorospheres and the tetraONE system. The NIAID DAIDS New Technologies Evaluation Group. Clin Diagn Lab Immunol 7(3):344–351
Schnizlein-Bick CT, Spritzler J et al (2000) Evaluation of TruCount absolute-count tubes for determining CD4 and CD8 cell numbers in human immunodeficiency virus-positive adults. Site investigators and the NIAID DAIDS New Technologies Evaluation Group. Clin Diagn Lab Immunol 7(3):336–343
Denny TN, Gelman R et al (2008) A North American multilaboratory study of CD4 counts using flow cytometric panLeukogating (PLG): a NIAID-DAIDS Immunology Quality Assessment Program Study. Cytometry B Clin Cytom 74(Suppl 1):S52–S64
Maecker HT, Rinfret A et al (2005) Standardization of cytokine flow cytometry assays. BMC Immunol 6:13
Sutherland DR, Anderson L et al (1996) The ISHAGE guidelines for CD34+ cell determination by flow cytometry. International Society of Hematotherapy and Graft Engineering. J Hematother 5(3):213–226
Perfetto SP, Chattopadhyay PK et al (2004) Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol 4(8):648–655
McLaughlin BE, Baumgarth N et al (2008) Nine-color flow cytometry for accurate measurement of T cell subsets and cytokine responses. Part II: panel performance across different instrument platforms. Cytometry A 73(5):411–420
Maecker HT, Frey T et al (2004) Selecting fluorochrome conjugates for maximum sensitivity. Cytometry A 62(2):169–173
Lamoreaux L, Roederer M et al (2006) Intracellular cytokine optimization and standard operating procedure. Nat Protoc 1(3):1507–1516
Horton H, Thomas EP et al (2007) Optimization and validation of an 8-color intracellular cytokine staining (ICS) assay to quantify antigen-specific T cells induced by vaccination. J Immunol Methods 323(1):39–54
Landay AL, Brambilla D et al (1995) Interlaboratory variability of CD8 subset measurements by flow cytometry and its applications to multicenter clinical trials. NAID/NICHD Women and Infants Transmission Study Group. Clin Diagn Lab Immunol 2(4):462–468
Levering WH, van Wieringen WN et al (2008) Flow cytometric lymphocyte subset enumeration: 10 years of external quality assessment in the Benelux countries. Cytometry B Clin Cytom 74(2):79–90
Britten CM, Gouttefangeas C et al (2008) The CIMT-monitoring panel: a two-step approach to harmonize the enumeration of antigen-specific CD8+ T lymphocytes by structural and functional assays. Cancer Immunol Immunother 57(3):289–302
Janetzki S, Panageas KS et al (2008) Results and harmonization guidelines from two large-scale international Elispot proficiency panels conducted by the Cancer Vaccine Consortium (CVC/SVI). Cancer Immunol Immunother 57(3):303–315
Britten CM, Janetzki S et al (2009) Harmonization guidelines for HLA-peptide multimer assays derived from results of a large scale international proficiency panel of the Cancer Vaccine Consortium. Cancer Immunol Immunother 58(10):1701–1713
Janetzki S, Price L et al (2010) Performance of serum-supplemented and serum-free media in IFNgamma Elispot Assays for human T cells. Cancer Immunol Immunother 59(4):609–618
Boedigheimer MJ, Ferbas J (2008) Mixture modeling approach to flow cytometry data. Cytometry A 73A(5):421–429
Chan C, Feng F et al (2008) Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry A 73(8):693–701
Lo K, Brinkman RR et al (2008) Automated gating of flow cytometry data via robust model-based clustering. Cytometry A 73(4):321–332
Pyne S, Hu X et al (2009) Automated high-dimensional flow cytometric data analysis. Proc Natl Acad Sci USA 106(21):8519–8524
Frelinger J, Kepler TB, Chan C (2008) Flow: statistics, visualization and informatics for flow cytometry. Source Code Biol Med 3:10. doi:10.1186/1751-0473-3-10
Titterington D, Smith AFM et al (1985) Statistical analysis of finite mixture distributions. Wiley, New York
Robert CP (1996) Mixtures of distributions: inference and estimation. Markov chain Monte Carlo in practice. Chapman & Hall, London, pp 441–464
Suchard M, Wang Q et al (2010) Understanding GPU programming for statistical computation: studies in massively parallel massive mixtures. J Comput Graph Stat. doi:10.1198/jcgs.2010.10016
Feyerabend S, Stevanovic S, Gouttefangeas C et al (2009) Novel multi-peptide vaccination in Hla-A2+ hormone sensitive patients with biochemical relapse of prostate cancer. Prostate 69:917–927
Ornatsky O, Baranov VI et al (2006) Multiple cellular antigen detection by ICP-MS. J Immunol Methods 308(1–2):68–76
Hadrup SR, Bakker AH et al (2009) Parallel detection of antigen-specific T-cell responses by multidimensional encoding of MHC multimers. Nat Methods 6(7):520–526
Newell EW, Klein LO et al (2009) Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat Methods 6(7):497–499
Acknowledgments
We gratefully acknowledge the many helpful discussions and collaboration with the statistics group led by Mike West (Duke University), as well as with the Cancer Immunotherapy Consortium (CIC) ImmunoAssay Working Group (IAWG), Cancer Immunotherapy Association (CIMT) Cancer Immunoguiding Program (CIP) and NIAID/BD ICS Quality Assurance Program (QAP) steering committees. Research supported by the National Institutes of Health (RC1AI086032-01, UL1RR024128 Cliburn Chan). The Center for Aids Research (CFAR) Flow Cytometry Core is supported by NIH grant 1P30 AI 64518.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is a Focussed Research Review based on a presentation given at the Seventh Annual Meeting of the Association for Immunotherapy of Cancer (CIMT), held in Mainz, Germany, 3–5 June 2009.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Frelinger, J., Ottinger, J., Gouttefangeas, C. et al. Modeling flow cytometry data for cancer vaccine immune monitoring. Cancer Immunol Immunother 59, 1435–1441 (2010). https://doi.org/10.1007/s00262-010-0883-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00262-010-0883-4