Abstract
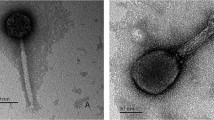
Bacteriophages infecting Staphylococcus epidermidis were isolated by mitomycin C induction. Three distinct phages (vB_SepiS-phiIPLA5, vB_SepiS-phiIPLA6, and vB_SepiS-phiIPLA7)—defined by plaque morphology, structure, virion proteins pattern, DNA restriction bands, and host range—were obtained. One-step growth curves of bacteriophages under optimal growth conditions for S. epidermidis F12 revealed eclipse and latent periods of 5–10 and 10–15 min, respectively, with burst sizes of about 5 to 30 PFU per infected cell. Transmission electron microscopy revealed that the phages were of similar size and belonged to the Siphoviridae family. Phage phi-IPLA7 had the broadest host range infecting 21 out of 65 S. epidermidis isolates. Phage phi-IPLA5 seemed to be a virulent phage probably derived from phi-IPLA6. Phages phi-IPLA5 and phi-IPLA7 exhibited increasing plaques surrounded by a halo that could be indicative of a polysaccharide depolymerase activity. Viable counts, determined during the infection of S. epidermidis F12, confirmed that phi-IPLA5 had a potent lytic capability and reduced S. epidermidis population by 5.67 log units in 8 h of incubation; in the presence of the mixture of phi-IPLA6 and phi-IPLA7, however, a reduction of 2.27 log units was detected




Similar content being viewed by others
References
Cairns B, Timms AR, Jansen VA, Connerton IF, Payne RJ (2009) Quantitative models of in vitro bacteriophage-host dynamics and their application to phage therapy. PLoS Pathog 5:e1000253
Carlson NG, Little JW (1993) A novel antivirulence element in the temperate bacteriophage HK022. J Bacteriol 175:7541–7549
Curtin JJ, Donlan RM (2006) Using bacteriophages to reduce formation of catheter-associated biofilms by Staphylococcus epidermidis. Antimicrob Agents Chemother 50:1268–1275
Daniel A, Bonnen PE, Fischetti VA (2007) First complete genome sequence of two Staphylococcus epidermidis bacteriophages. J Bacteriol 189:2086–2100
Delgado S, Arroyo R, Jimenez E, Marin ML, Del Campo R, Fernandez L, Rodriguez JM (2009) Staphylococcus epidermidis strains isolated from breast milk of women suffering infectious mastitis: potential virulence traits and resistance to antibiotics. BMC Microbiol 9:82
García P, Ladero V, Suárez JE (2003) Analysis of the morphogenetic cluster and genome of the temperate Lactobacillus casei bacteriophage A2. Arch Virol 148:1–20
García P, Madera C, Martínez B, Rodríguez A, Suárez JE (2009) Prevalence of bacteriophages infecting Staphylococcus aureus in dairy samples and their potential as biocontrol agents. J Dairy Sci 92:3019–3026
Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438
Herrero M, de los Reyes-Gavilán CG, Caso JL, Suárez JE (1994) Characterization of ϕ393-A2, a bacteriophage that infects Lactobacillus casei. Microbiology 140:2585–2590
Hughes KA, Sutherland IW, Clark J, Jones MV (1998) Bacteriophage and associated polysaccharide depolymerases - novel tools for study of bacterial biofilms. J Appl Microbiol 85:583–590
Laemmli U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
McCann MT, Gilmore BF, Gorman SP (2008) Staphylococcus epidermidis device-related infections: pathogenesis and clinical management. J Pharm Pharmacol 60:1551–1571
Merabishvili M, Pirnay JP, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R, Volckaert G, Mattheus W, Verween G, De Corte P, Rose T, Jennes S, Zizi M, De Vos D, Vaneechoutte M (2009) Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944
Moak M, Molineux IJ (2004) Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol Microbiol 51:1169–1183
Oliveira M, Nunes SF, Carneiro C, Bexiga R, Bernardo F, Vilela CL (2007) Time course of biofilm formation by Staphylococcus aureus and Staphylococcus epidermidis mastitis isolates. Vet Microbiol 124:187–191
Otto M (2008) Staphylococcal biofilms. Curr Top Microbiol Immunol 322:207–228
Otto M (2009) Staphylococcus epidermidis—the “accidental” pathogen. Nat Rev 7:555–567
Piette A, Verschraegen G (2009) Role of coagulase-negative staphylococci in human disease. Vet Microbiol 134:45–54
Sambrook J, Maniatis T, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sulakvelidze A, Kutter E (2005) Bacteriophage therapy in humans. In: Kutter E, Sulakvelidze A (eds) Bacteriophages: biology and application. CRC Press, Boca Raton, pp 381–436
Talbot HW Jr, Parisi JT (1976) Phage typing of Staphylococcus epidermidis. J Clin Microbiol 3:519–523
Wright A, Hawkins CH, Anggård EE, Harper DR (2009) A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin Otolaryngol 34:349–357
Acknowledgments
This research study was supported by grants AGL2009-13144-C02-01 from the Ministry of Education of Spain and IB08-052 from FICYT (Regional Government of Asturias). P.G. was a fellow of the Spanish Ministry of Education Ramón y Cajal Research Programme. We thank Dr. J. M. Rodríguez (Fac. Veterinaria, UCM, Madrid) for providing the S. epidermidis strains.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gutiérrez, D., Martínez, B., Rodríguez, A. et al. Isolation and Characterization of Bacteriophages Infecting Staphylococcus epidermidis . Curr Microbiol 61, 601–608 (2010). https://doi.org/10.1007/s00284-010-9659-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9659-5