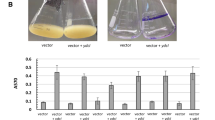
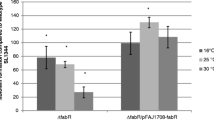
Abstract
Two lineages of enterohemorrhagic Escherichia coli O157:H7 (EDL933, Stx1+ and Stx2+) and 86-24 (Stx2+) were investigated to determine the genetic basis of biofilm formation on abiotic surfaces. Strain EDL933 formed a robust biofilm while strain 86-24 formed almost no biofilm on either polystyrene plates or polyethylene tubes. Whole-transcriptome profiles of EDL933 versus 86-24 revealed that in the strong biofilm-forming strain, genes involved in curli biosynthesis and cellulose production were significantly induced, whereas genes involved in indole signaling were most repressed. Additionally, 49 phage genes were highly induced and repressed between the two strains. Curli assays using Congo red plates and scanning electron microscopy corroborated the microarray data as the EDL933 strain produced a large amount of curli, while strain 86-24 formed much less curli. Moreover, EDL933 produced 19-fold more cellulose than 86-24, and indole production in EDL933 was two times lower than that of the strain 86-24. Therefore, it appears E. coli O157:H7 EDL933 produces more biofilm because of its increased curli and cellulose production and reduced indole production.


Similar content being viewed by others
References
Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A (2007) Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun 75:4597–4607
Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A (2008) Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol 78:811–819
Bokranz W, Wang X, Tschäpe H, Römling U (2005) Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol 54:1171–1182
Boyce TG, Swerdlow DL, Griffin PM (1995) Escherichia coli O157:H7 and the hemolytic-uremic syndrome. N Engl J Med 333:364–368
Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ (2008) The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27
Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ (2002) Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855
Cornick NA, Helgerson AF, Sharma V (2007) Shiga toxin and Shiga toxin-encoding phage do not facilitate Escherichia coli O157:H7 colonization in sheep. Appl Environ Microbiol 73:344–346
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322
Dowd SE, Ishizaki H (2006) Microarray based comparison of two Escherichia coli O157:H7 lineages. BMC Microbiol 6:30
Frenzen PD, Drake A, Angulo FJ (2005) Economic cost of illness due to Escherichia coli O157 infections in the United States. J Food Prot 68:2623–2630
Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA (1988) Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med 109:705–712
Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Høiby N, Kjelleberg S, Givskov M (2002) Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148:87–102
Herold S, Siebert J, Huber A, Schmidt H (2005) Global expression of prophage genes in Escherichia coli O157:H7 strain EDL933 in response to norfloxacin. Antimicrob Agents Chemother 49:931–944
Jarvis KG, Kaper JB (1996) Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun 64:4826–4829
Kawamura-Sato K, Shibayama K, Horii T, Iimuma Y, Arakawa Y, Ohta M (1999) Role of multiple efflux pumps in Escherichia coli in indole expulsion. FEMS Microbiol Lett 179:345–352
Kimmitt PT, Harwood CR, Barer MR (2000) Toxin gene expression by shiga toxin-producing Escherichia coli: the role of antibiotics and the bacterial SOS response. Emerg Infect Dis 6:458–465
Lazazzera BA (2005) Lessons from DNA microarray analysis: the gene expression profile of biofilms. Curr Opin Microbiol 8:222–227
Lee J-H, Lee J (2010) Indole as an intercellular signal in microbial community. FEMS Microbiol Rev 34:426–444
Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK (2007) Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol 73:4100–4109
Lee J, Zhang XS, Hegde M, Bentley WE, Jayaraman A, Wood TK (2008) Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J 2:1007–1023
Lee Y, Kim Y, Yeom S, Kim S, Park S, Jeon CO, Park W (2008) The role of disulfide bond isomerase A (DsbA) of Escherichia coli O157:H7 in biofilm formation and virulence. FEMS Microbiol Lett 278:213–222
Lee J-H, Cho MH, Lee J (2011) 3-Indolylacetonitrile decreases Escherichia coli O157:H7 biofilm formation and Pseudomonas aeruginosa virulence. Environ Microbiol 13:62–73
Lesic B, Lépine F, Déziel E, Zhang J, Zhang Q, Padfield K, Castonguay MH, Milot S, Stachel S, Tzika AA, Tompkins RG, Rahme LG (2007) Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog 3:1229–1239
Ma Q, Wood TK (2009) OmpA influences Escherichia coli biofilm formation by repressing cellulose production through the CpxRA two-component system. Environ Microbiol 11:2735–2746
Matthysse AG, Deora R, Mishra M, Torres AG (2008) Polysaccharides cellulose, poly-beta-1, 6-N-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74:2384–2390
Nataro JP, Kaper JB (1998) Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201
Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533
Potera C (1999) Forging a link between biofilms and disease. Science 283:1837–1839
Pratt LA, Kolter R (1998) Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30:285–293
Prigent-Combaret C, Prensier G, Le Thi TT, Vidal O, Lejeune P, Dorel C (2000) Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ Microbiol 2:450–464
Puttamreddy S, Cornick NA, Minion FC (2010) Genome-wide transposon mutagenesis reveals a role for pO157 genes in biofilm development in Escherichia coli O157:H7 EDL933. Infect Immun 78:2377–2384
Reisner A, Krogfelt KA, Klein BM, Zechner EL, Molin S (2006) In vitro biofilm formation of commensal and pathogenic Escherichia coli strains: impact of environmental and genetic factors. J Bacteriol 188:3572–3581
Ren D, Bedzyk LA, RW Ye, Thomas SM, Wood TK (2004) Differential gene expression shows natural brominated furanones interfere with the autoinducer-2 bacterial signaling system of Escherichia coli. Biotechnol Bioeng 88:630–642
Rivas L, Dykes GA, Fegan N (2007) A comparative study of biofilm formation by Shiga toxigenic Escherichia coli using epifluorescence microscopy on stainless steel and a microtitre plate method. J Microbiol Meth 69:44–51
Ryu JH, Beuchat LR (2005) Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl Environ Microbiol 71:247–254
Saldaña Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Girón JA (2009) Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ Microbiol 11:992–1006
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O’Brien AD (1986) Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53:135–140
Tarr PI, Gordon CA, Chandler WL (2005) Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086
Uhlich GA, Cooke PH, Solomon EB (2006) Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl Environ Microbiol 72:2564–2572
Uhlich GA, Gunther NWt, Bayles DO, Mosier DA (2009) The CsgA and Lpp proteins of an Escherichia coli O157:H7 strain affect HEp-2 cell invasion, motility, and biofilm formation. Infect Immun 77:1543–1552
Wang X, Rochon M, Lamprokostopoulou A, Lünsdorf H, Nimtz M, Römling U (2006) Impact of biofilm matrix components on interaction of commensal Escherichia coli with the gastrointestinal cell line HT-29. Cell Mol Life Sci 63:2352–2363
Wang X, Kim Y, Wood TK (2009) Control and benefits of CP4-57 prophage excision in Escherichia coli biofilms. ISME J 3:1164–1179
Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK (1983) Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol 18:512–520
Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA (2008) EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604
Yoon Y, Sofos JN (2008) Autoinducer-2 activity of gram-negative foodborne pathogenic bacteria and its influence on biofilm formation. J Food Sci 73:M140–M147
Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U (2001) The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39:1452–1463
Acknowledgments
This research was supported by the Yeungnam University research grant in 2009, and the Human Resources Development Program (R&D Workforce Cultivation Track for Solar Cell Materials and Processes) of Korea Institute of Energy Technology Evaluation and Planning (KETEP) grant (No. 20104010100580) funded by the Korean Ministry of Knowledge Economy. T. Wood is the T. Michael O’Connor endowed chair and is also supported by the NIH (R01 GM089999).
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, JH., Kim, YG., Cho, M.H. et al. Transcriptomic Analysis for Genetic Mechanisms of the Factors Related to Biofilm Formation in Escherichia coli O157:H7. Curr Microbiol 62, 1321–1330 (2011). https://doi.org/10.1007/s00284-010-9862-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-010-9862-4