Abstract
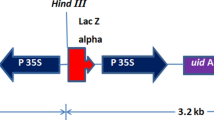
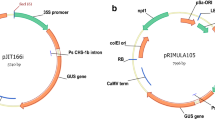
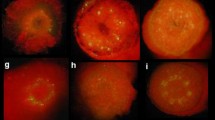
We have developed a high-throughput Agrobacterium-mediated transformation model system using both nptII and the 5-enolpyruvylshikimate-3-phosphate synthase gene from Agrobacterium tumefaciens strain CP4 (cp4) based selections in MicroTom, a miniature rapid-cycling cherry tomato variety. With the NPTII selection system, transformation frequency calculated as independent transgenic events per inoculated explant ranged from 24 to 80% with an average of 56%, in industrial production scale transformation experiments. For CP4, with glyphosate selection, the average transformation frequency was 57%. Stable transformation frequency was positively correlated with transient expression (R=0.85), and variable with the genes of interest. DNA integration and germline transformation were confirmed by biological assay, Southern Blot analysis, and R1 phenotype segregation. Transgene expression was observed in leaf, root, stem, flower, and fruit tissues of the transgenic plants. Ninety-five percent of transgenic events coexpressed two introduced genes based on β-glucuronidase (GUS) and neonmycin phosphotransferase II (NPTII) expression. Seventy-five percent of transgenic events contained one to two copies of the introduced uidA (GUS) gene based on Southern analysis. Transgenic plants from the cotyledon explants to the transgenic plants transferred to soil were produced within about 2–3 months depending on the genes of interest. The utility of this MicroTom model transformation system for functional genomic studies, such as identification of genes related to important agricultural traits and gene function, is discussed.






Similar content being viewed by others
References
Atherton JG, Rudich J (1986) The tomato Crop. Chapman and Hall, London, New York, 661 pp
Barry G, Kishore G, Padgette S, Taylor M, Kolacz K, Weldon M, Re D, Eichholtz D, Fincher D, Hallas L (1992) Inhibitors of amino acid biosynthesis: strategies for imparting glyphosate tolerance to crop plants. In: Sinch BK, Flores HE, Shannon JC (eds) Biosynthesis and molecular regulation of amino acids in plants. American Society of Plant Physiologists, Rochville, MD, pp 139–145
Chen LZ, Adachi T (1994) Plant regeneration via somatic embryogenesis from cotyledon protoplasts of tomato (Lycopersicon esculentum). Breed Sci 44:257–262
Dan YH, Yan H, Munyikwa T, Dong J, Zhang B, Lahman L, Rommens C (2001) MicroTom—a model functional genomics assay. In Vitro Cell Dev Biol Plant 37:22-A
FAS/USDA (2003) Processed tomato products outlook and situation in selected counties. Horticutural and Tropical Products Division
Fillatti J, Kiser J, Rose R, Comai L (1987) Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Biotechnology 5:726–730
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirement of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hinchee MAW, Corbin DR, Armstrong CL, Fry JE, Sato SS, DeBoer DL, Petersen WL, Armstrong TA, Connor-Ward DV, Layton JG, Horsch RB (1994) Plant transformation. In: Vasil IK, Thorpe TA (eds) Plant cell and tissue culture. Academic, Dordrecht, The Netherlands, pp 231–270
Horsch RB, Fry J, Hoffmann N, Wallroth M, Eichholtz D, Rogers S, Fraley R (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Horsch R, Fraley R, Rogers S, Fry J, Klee H, Shah D, McCormick S, Niedermeyer J, Hoffmann N (1987) Agrobacterium-mediated transformation of plants. In: Green CE, Somers DA, Hackett WP, Biesboer DD (eds) Plant tissue and cell culture. Geo J Ball, West Chicago, pp 317–329
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Lee S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G (2000) T-DNA insertional mutagenesis for functional genomics in rice. Plant J 22:561–570
Jiang GY, Jin DM, Weng ML, Guo BT, Wang B (1999) Transformation and expression of trichosanthin gene in tomato. Acta Bot Sin 41:334–336
Jones DA, Thomas CM, Hammond-Kossack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266:789–793
Kay R, Chan A, Daly M, McPherson J (1987) Duplication of CaMV 35S promoter sequences creates a strong enhancer for plant genes. Science 230:1299–1302
Koornneef M, Hanhart C, Jongsma M, Toma I, Weide R, Zabel P, Hille J (1986) Breeding of a tomato genotype readily accessible to genetic manipulation. Plant Sci 45:201–208
Krasnyanski S, Sandhu J, Domier LL, Buetow DE, Korban SS (2001) Effect of enhanced CAMV 35S promoter and a fruit-specific promoter on uida gene expression in transgenic tomato plants. In Vitro Cell Dev Biol Plant 37:427–433
Lagudah ES, Dubcovsky J, Powell W (2001) Wheat genomics. Plant Physiol Biochem 39:335–344
Lee MK, Kim HS, Kim JS, Kim SH, Park YD (2004) Agrobacterium-mediated transformation system for large-scale production of transgenic Chinese cabbage (Brassica rapa L. ssp pekinensis) plants for insertional mutagenesis. J Plant Biol 47:300–306
Mathews H, Clendennen KS, Caldwell GC, Liu XL, Connors K, Matheis N, Schuster KD, Menasco DJ, Wagoner W, Lightner J, Wagner DR (2003) Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 15:1689–1703
McCormick S, Niedermeyer J, Fry J, Barnason A, Horsch R, Fraley R (1986) Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep 5:81–84
Meinke DW, Cherry JM, Dean C, Rounsley SD, Koornneef M (1998) Arabidopsis thaliana: a model plant for genome analysis. Science 282:679–682
Meissner R, Jacobson Y, Melame S, Levyatuv S, Shalev G, Ashri A, Elkind Y, Levy A (1997) A new model system for tomato genetics. Plant J 12:1465–1472
Meyerowitz E, Sommerville CR (1994) Arabidopsis. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Olhoft PM, Flagel LE, Somers DA (2004) T-DNA locus structure in a large population of soybean plants transformed using the Agrobacterium-mediated cotyledonary-node method. Plant Biotechnol J 2:289–300
Ostergaard L, Yanofsky MF (2004) Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J 39:682–696
Pereira A (2000) A transgenic perspective on plant functional genomics. Transgenic Res 9:245–260
Rogers SG (1990) Promoter for transgenic plants. US Patent No. 05378619
Rommens C (2000) Discovery of new signaling genes associated with late blight resistance in patato. Cambridge Healthtech Institute's Third Annual Impact of Molecular Biology on Crop Production and Crop Protection (abstract)
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Scott JW, Harbaugh BK (1989) MicroTom—a miniature dwarf tomato. Fla Agr Exp Sta Circ 370:1–6
Shen L, Sheng JP, Luo YB (1998) Tomato transgenic system with Ti plasmid of Agrobacterium tumefaciens. J China Agric Univ 3:101–110
Shibata D (2005) Genome sequencing and functional genomics approaches in tomato. J Gen Plant Pathol 71:1–7
Sidorova N, Layton JG, Armstrong CL (2001) Comparison of model transformation systems. In Vitro Cell Dev Biol Plant 37:20-A
Takahashi H, Shimizu A, Arie T, Rosmalawati S, Fukushima S, Kikuchi M, Hikichi Y, Kanda A, Takahashi A, Kiba A, Ohnishi K, Ichinose Y, Taguchi F, Yasuda C, Kodama M, Egusa M, Masuta C, Sawada H, Shibata D, Hori K, Watanabe Y (2005) Catalog of Micro-Tom tomato responses to common fungal, bacterial, and viral pathogens. J Gen Plant Pathol 71:8–22
Tsugane T, Watanabe M, Yano K, Sakurai N, Suzuki H, Shibata D (2005) Expressed sequence tags of full-length cDNA clones from the miniature tomato (Lycopersicon esculentum) cultivar Micro-Tom. Plant Biotechnol 22:161–165
Tyagi AK, Mohanty A (2000) Rice transformation for crop improvement and functional genomics. Plant Sci 158:1–18
Webb RE, Stoner AK, Gentile AG (1973) Tomatoes. In: Nelson RR (ed) Plants for disease resistance breeding. Pennsylvania State University Press, University Park, pp 344–360
Wei H, Phillips GC (2001) A combination of overgrowth-control antibiotics improves Agrobacterium tumefaciens-mediated transformation efficiency for cultivated tomato (L. esculentum). In Vitro Cell Dev Biol Plant 37:12–18
Yamamoto N, Tsugane T, Watanabe M, Yano K, Maeda F, Kuwata C, Torki M, Ban Y, Nishimura S, Shibata D (2005) Expressed sequence tags from the laboratory-grown miniature tomato (Lycopersicon esculentum) cultivar Micro-Tom and mining for single nucleotide polymorphisms and insertions/deletions in tomato cultivars. Gene 356:127–134
Yang YZ, Peng H, Huang HM, Wu JX, Ha SR, Huang DF, Lu TG (2004) Large-scale production of enhancer trapping lines for rice functional genomics. Plant Sci 167:281–288
Acknowledgements
The authors would like to thank David Duncan for critically reviewing the manuscript and for helpful suggestions. The authors are grateful to Jeanne Layton and Pamela Delaquil for providing us technical information on standard tomato transformation at Monsanto. The authors would like to thank Linda Lahman, Salim Hakimi, Glenda Debrecht, and Bei Zhang for their technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. C. Jordan
Rights and permissions
About this article
Cite this article
Dan, Y., Yan, H., Munyikwa, T. et al. MicroTom—a high-throughput model transformation system for functional genomics. Plant Cell Rep 25, 432–441 (2006). https://doi.org/10.1007/s00299-005-0084-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-005-0084-3