Abstract
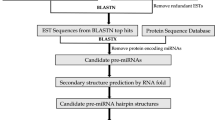
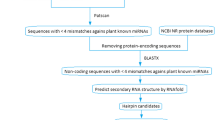
Micro-RNAs (miRNA) are regulatory non-coding class of small RNAs functioning in many organisms. Using computational approaches we have identified 20 conserved opium poppy (Papaver somniferum L.) miRNAs belonging to 16 miRNA families in Expressed Sequence Tags (EST) database. The existence of ESTs suggested that the miRNAs were expressed in P. somniferum. Lengths of mature miRNAs varied from 20 to 23 nucleotides located at the different positions of precursor RNAs. Uracil was found to be a dominant nucleotide in both poppy pre-miRNA sequences (31.28 ± 7.06% of total nucleotide composition) and in the first position at the 5′ end of the mature poppy miRNAs. We have applied quantitative real-time PCR (qRT-PCR) assays to compare and validate expression levels of selected P. somniferum miRNAs and their target transcripts. As a result, some of the predicted miRNAs and their target genes were found to be differentially expressed in P. somniferum leaf and root tissues. A meaningful correlation between three of the four analyzed pairs of miRNAs and their target transcript expression levels was detected. Additionally, using these predicted miRNAs as queries, 41 potential target mRNAs were found in National Center for Biotechnology Information (NCBI) protein-coding nucleotide (mRNA) database of all plant species. Some of the target mRNAs were found to be transcription factors regulating plant development, morphology, and flowering time. Other target mRNAs of identified conserved miRNAs involve in metabolic processes, signal transduction, and stress responses. This study reports the first identification of opium poppy miRNAs.




Similar content being viewed by others
References
Achard P, Herr A, Baulcombe DC, Harberd N (2004) Modulation of floral development by a gibberellin-regulated microRNA. Development 131:3357–3365
Allen R, Millgate AG, Chitty JA, Thisleton J, Miller JAC, Fist AJ, Gerlach WL, Larkin PJ (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566
Altschul S, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Aukerman M, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Bao N, Lye KW, Barton MK (2004) MicroRNA binding sites in Arabidopsis class IIIHD-ZIP mRNAs are required for methylation of the template chromosome. Dev Cell 7:653–662
Bartel D (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Detection of 91 potential in plant conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci USA 101:11511–11516
Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320:1185–1190
Cai X, Ballif J, Endo S, Davis E, Liang M, Chen D, DeWald D, Kreps J, Zhu T, Wu YA (2007) Putative CCAAT-binding transcription factor is a regulator of flowering timing in Arabidopsis. Plant Physiol 145:98–105
Carrington J, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen X (2005) MicroRNA biogenesis and function in plants. FEBS Lett 579:5923–5931
Chen J, Li WX, Xie D, Peng JR, Ding SW (2004) Viral virulence protein suppresses RNA silencing-mediated defense but upregulates the role of microRNA in host gene expressio. Plant Cell 16:1302–1313
Guo Q, Xiang AL, Yang Q, Yang ZM (2007) Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull 52:2380–2389
Jones-Rhoades M, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jung J, Park CM (2007) MIR166/165 genes exhibit dynamic expression patterns in regulating leaf apical meristem and floral development in Arabidopsis. Planta 225:1327–1338
Kasschau K, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4:205–217
Kim J, Jung JH, Reyes JL, Kim YS, Kim SY, Chung KS, Kim JA, Lee M, Lee Y, Kim VN, Chua NH, Park CM (2005) microRNA-directed cleavage of ATHB15 mRNA regulates vascular development in Arabidopsis inflorescence stems. Plant J 42:84–94
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101:12753–12758
Laufs P, Peaucelle A, Morin H, Traas J (2004) MicroRNA regulation of theCUC genes is required for boundary size control in Arabidopsis meristems. Dev Cell 131:4311–4322
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci USA 102:9412–9417
Lin S, Chang D, Ying SY (2005) Asymmetry of intronic pre-miRNA structures in functional RISC assembly. Gene 356:32–38
Llave C, Xie ZX, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Luo Y, Zhou H, Li Y, Chen JY, Yang JH, Chen YQ, Qu LH (2006) Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Lett 580:5111–5116
Mallory A, Dugas DV, Bartel DP, Bartel B (2004) MicroRNA regulation of NAC-domain targets is required for proper formation and separation of adjacent embryonic, vegetative, and floral organs. Curr Biol 14:1035–1046
Millar A, Gubler F (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Cell 17:705–721
Mlotshwa S, Yang Z, Kim Y, Chen X (2006) Floral patterning defects induced by Arabidopsis APETALA2 and micro- RNA172 expression in Nicotiana benthamiana. Plant Mol Biol 61:781–793
Ochando I, Jover-Gil S, Ripoll JJ, Candela H, Vera A, Ponce MR, Martínez-Laborda A, Micol JL (2006) Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in Arabidopsis. Plant Physiol 14:607–619
Page E (2005) Silencing nature’s narcotics: metabolic engineering of the opium poppy. Trends Biotechnol 23:331–332
Palatnik J, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D (2003) Control of leaf morphogenesis by microRNAs. Nature 425:257–263
Park W, Li JJ, Song RT, Messing J, Chen XM (2002) CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 12:1484–1495
Prigge M, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17:61–76
Reinhart B, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Gene Dev 16:1616–1626
Rhoades M, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110:513–520
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P (2008) The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol 67:183–195
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16:2001–2019
Udvardi MK, Czechowski T, Scheible W-R (2008) Eleven golden rules of quantitative RT-PCR. Plant Cell 20:1736–1737
Unver T, Budak H (2009) Conserved microRNAs and their targets in model grass species Brachypodium distachyon. Planta 230:659–669
Unver T, Bozkurt O, Akkaya MS (2008) Identification of differentially expressed transcripts from leaves of the boron tolerant plant Gypsophila perfoliata L. Plant Cell Rep 27:1411–1422
Unver T, Namuth-Covert D, Budak H (2009) Review of current methodological approaches for characterizing microRNAs in plants Int J Plant Genom 262–463
Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Method 3:1–12
Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166 g and its AtHDZIP target genes. Development 132:3657–3668
Xie F, Huang SQ, Guo K, Xiang AL, Zhu YY, Nie L, Yang ZM (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581:1464–1474
Yin Z, Li C, Han X, Shen F (2008) Identification of conserved microRNAs and their target genes in tomato (Lycopersicon esculentum). Gene 414:60–66
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 33(Web Server issue):W701–4
Zhang B, Pan XP, Wang QL, Cobb GP, Anderson TA (2005) Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15:336–360
Zhang B, Pan XP, Anderson TA (2006a) Identification of 188 conserved maize microRNAs and their targets. FEBS Lett 580:3753–3762
Zhang B, Pan XP, Cobb GP, Anderson TA (2006b) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289:3–16
Zhang B, Pan XP, Wang QL, Cobb GP, Anderson TA (2006c) Computational identification of microRNAs and their targets. Comput Biol Chem 30:395–407
Zhang B, Wang QL, Pan XP (2007a) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210:279–289
Zhang B, Wang QL, Wang K, Pan X, Liu F, Gou T, Cobb GP, AT A (2007b) Identification of cotton microRNAs and their targets. Gene 397:26–37
Zhang B, Pan X, Stellwag EJ (2008) Identification of soybean microRNAs and their targets. Planta 229:161–182
Zhang W, Luo Y, Gong X, Zeng W, Li S (2009) Computational identification of 48 potato microRNAs and their targets. Comput Biol Chem 33:84–93
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgments
We would like to thank Prof. Neset Arslan of Ankara University for supplying poppy seeds and Prof. Hikmet Budak of Sabancı University for his kind assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Judelson.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Unver, T., Parmaksız, İ. & Dündar, E. Identification of conserved micro-RNAs and their target transcripts in opium poppy (Papaver somniferum L.). Plant Cell Rep 29, 757–769 (2010). https://doi.org/10.1007/s00299-010-0862-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-010-0862-4