Abstract
PICH (Plk1-interacting checkpoint helicase) was recently identified as an essential component of the spindle assembly checkpoint and shown to localize to kinetochores, inner centromeres, and thin threads connecting separating chromosomes even during anaphase. In this paper, we have used immuno-fiber fluorescence in situ hybridization and chromatin-immunoprecipitation to demonstrate that PICH associates with centromeric chromatin during anaphase. Furthermore, by careful analysis of PICH-positive anaphase threads through FISH as well as bromo-deoxyurdine and CREST labeling, we strengthen the evidence that these threads comprise mainly alphoid centromere deoxyribonucleic acid. Finally, by timing the addition of ICRF-193 (a specific inhibitor of topoisomerase-II alpha) to cells synchronized in anaphase, we demonstrate that topoisomerase activity is required specifically to resolve PICH-positive threads during anaphase (as opposed to being required to prevent the formation of such threads during earlier cell cycle stages). These data indicate that PICH associates with centromeres during anaphase and that most PICH-positive threads evolve from inner centromeres as these stretch in response to tension. Moreover, they show that topoisomerase activity is required during anaphase for the resolution of PICH-positive threads, implying that the complete separation of sister chromatids occurs later than previously assumed.
Similar content being viewed by others
Introduction
The SNF2 family ATPase PICH was recently discovered as an essential component of the spindle assembly checkpoint (SAC; Baumann et al. 2007). This surveillance mechanism monitors the bipolar attachment of chromosomes to the mitotic spindle and is crucial for correct segregation of genetic information during cell division (Chan et al. 2005; Kops et al. 2005; Musacchio and Salmon 2007). The SAC is thought to be regulated by either the tension that develops between sister chromatids in response to bipolar attachment or, alternatively, the increased microtubule occupancy at kinetochores that results from the tension-induced stabilization of kinetochore–microtubule interactions. It is interesting to note that PICH localizes to kinetochores and inner centromeres, prompting the hypothesis that it may function as a tension sensor in SAC signaling (Baumann et al. 2007). Unexpectedly, PICH also decorated ultrathin threads that appeared to connect the kinetochores of sister chromatids even after anaphase onset. As cells progressed through anaphase, PICH-positive threads became progressively longer and, concomitantly, their numbers diminished so that they were no longer detectable by telophase (Baumann et al. 2007). Although these threads were sensitive to DNase I, they could not be counterstained with conventional deoxyribonucleic acid (DNA) dyes or antibodies against histones (Baumann et al. 2007; Chan et al. 2007). It is interesting to note that, however, Bloom’s syndrome protein (BLM), along with its cellular partners, topoisomerase IIIα (TopoIIIα) and hRMI1, was recently found to associate with PICH-positive threads (Chan et al. 2007). As the BLM/TopoIIIα/hRMI1 complex is required for the dissolution of recombination and/or replication intermediates (Wu and Hickson 2003; Seki et al. 2006; Wu et al. 2006), it was proposed that PICH-BLM-positive DNA structures may originate from incompletely replicated DNA or fully replicated but still intertwined (catenated) duplexes (Chan et al. 2007).
Sister chromatid cohesion is established during S phase and relies on two mechanisms. First, catenation of sister chromatids is an inevitable consequence of DNA replication, so that topoisomerase II (Topo-II) activity is required for DNA decatenation and sister chromatid separation (Sundin and Varshavsky 1981; Clarke et al. 1993; Yanagida 2005; Toyoda and Yanagida 2006). Second, proteins known as cohesins are loaded onto DNA and proposed to form ring-like structures embracing the two 10-nm sister chromatid fibers (Nasmyth and Haering 2005; Losada 2007). In vertebrates, cohesin proteins are removed from chromosome arms already during prophase, in response to the action of the protein Wapl (Gandhi et al. 2006; Kueng et al. 2006) and to phosphorylation of a cohesin subunit by Plk1 and Aurora B (Losada et al. 2002; Sumara et al. 2002). At centromeres, cohesin proteins persist, partly because of the protective action of a complex between shugoshin 1 and protein phosphatase 2A (Kitajima et al. 2006; Riedel et al. 2006; Tang et al. 2006). This centromeric pool of cohesin is only removed after silencing of the SAC, when activation of the anaphase-promoting complex/cyclosome results in the release of the protease separase, which then cleaves the cohesin subunit hRad21/Scc1 (Nasmyth and Haering 2005).
The path of sister chromatid DNA at centromeres and the timing of decatenation of these late-replicating structures remains poorly understood (Yanagida 2005; Bloom et al. 2006; Baumann et al. 2007). At one time, DNA catenation had been considered a potentially important mechanism for sister chromatid cohesion (Murray and Szostak 1985). However, after studies with yeast minichromosomes argued against this model (Koshland and Hartwell 1987), most of the more recent work was focused primarily on the role of cohesin proteins. Yet, Topo-II is clearly required for chromosome segregation (Toyoda and Yanagida 2006). Moreover, one biochemical study performed in Xenopus egg extracts clearly showed a requirement for Topo-II activity after the onset of anaphase (Shamu and Murray 1992), raising the possibility that centromeres are still catenated while sister chromatids undergo bipolar attachment. If this were the case, it would imply a structural continuity between centromeric sister chromatid DNA at the metaphase to anaphase transition. This has potentially important implications with regard to the question of where tension might be monitored by the SAC. If sister chromatid cohesion were mediated exclusively by centromere-associated cohesins, then these proteins would represent the only device for opposing the pulling forces of kinetochore microtubules. However, if centromere DNA persisted in a catenated state, then this DNA would be stretched in response to bipolar attachment, raising the possibility that developing tension could be monitored through a DNA-binding protein located on catenated centromeres (Baumann et al. 2007).
Interest in the path of centromere DNA and the timing of centromere decatenation has been renewed by the recent identification of PICH as a candidate tension sensor and the discovery of PICH-positive threads in anaphase cells (Baumann et al. 2007). Following up on these findings, we have here addressed two important issues. First, in order for PICH to function as a tension sensor according to the proposed model, the protein was predicted to associate preferentially with centromeric chromatin. Second, the discovery of PICH-positive threads in anaphase cells suggested that decatenation of centromeric chromatin might be completed later than hitherto assumed. These predictions were tested, first, by the application of fluorescence in situ hybridization (FISH) and antibody probes to PICH-positive DNA structures and the use of complementary chromatin immunoprecipitation (ChIP) and, second, careful studies on the role of topoisomerase-IIα (Topo-IIα) in the resolution of PICH-positive threads specifically during anaphase. Our data clearly demonstrate that PICH associates with centromeric chromatin in anaphase cells and that the resolution of PICH-positive threads requires Topo-II activity after anaphase onset. These results have important implications for the path of centromere DNA and the timing of sister chromatid separation. Specifically, they indicate that sister chromatid centromeres in human cells are held together not only through protein-mediated cohesion but also through catenation and that complete decatenation of centromere DNA occurs only after anaphase onset.
Materials and methods
Preparation of anaphase cells and mechanically extended chromatin fibers
Immuno-fiber FISH was used for detecting specific DNA sequences on extended chromatin fibers from anaphase cells. For labeling DNA with bromo-deoxyurdine (BrdU), cells were incubated with BrdU (BrdU-labeling reagent, diluted 1/500 in normal medium; Zymed Laboratories, South San Francisco, CA) for 10 h before Noscapine treatment. To enrich for anaphase cells, HeLaS3 cells were arrested in mitosis by 20 μM Noscapine for 18 h and then released into normal medium for 30 min. Anaphase cells were collected by mitotic shake-off, spun down (1,000 rpm, 5 min), and resuspended for 10 min into 75 mM KCl at a concentration of 2 × 106 cells/ml. Fifty microliters of cells were then spotted on positively charged microscope slides (SuperFrost Ultra Plus; Menzel Gläser), air dried, and immersed for 15 min at room temperature (RT) in salt detergent buffer (25 mM Tris pH 7.5, 500 mM NaCl, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]). Slides were then removed slowly from this solution and placed for 10 min into fixation buffer as described above. Subsequently, slides were washed twice in phosphate-buffered saline (PBS), 0.1% Twin-20, incubated for 2 min in 0.1 N HCl, rinsed twice in 2× sodium chloride–sodium citrate (SSC), and then used for FISH, followed by immunostaining with antibodies, as described below.
FISH probes
Biotin-labeled DNA probes for alphoid pan-centromere DNA, ribosomal DNA (rDNA), and telomeric DNA were prepared as described previously (Ijdo et al. 1991; Dunham et al. 1992; Langer et al. 2001).
Antibodies
Antibodies against PICH, Hec1, and CREST antiserum have been described (Baumann et al. 2007). Rabbit anti histone H3 (ab1791), H3 tri-methyl K9 (3mH3K9, ab8898), H3 di-methyl K4 (2mH3K4, ab7766), H3 acetyl K18 (ab1191), as well as mouse monoclonal anti-CENP-A (ab13939) were obtained from Abcam (Cambridge, UK). Mouse anti-BrdU (RPN20AB) was from BD Biosciences Pharmingen, NJ, and mouse monoclonal anti-TRF2 (4A794) from Upstate (Lake Placid, NY). The FISH secondary antibodies avidine-Cy3.5 was from Rockland Immunochemicals, Gilbertsville, PA. The following secondary antibodies were used in immunofluorescence (IF) staining: donkey anti-rat Cy3 and Cy2 conjugated (Jackson ImmunoResearch, West Grove, PA), Alexa Fluor 488 conjugated donkey anti-mouse or anti-rabbit, and Alexa Fluor 647 conjugated donkey anti-rabbit IgG (Molecular Probes, Invitrogen).
Preparation of cells for immunofluorescence and in situ hybridization
To achieve staining with both antibodies and DNA probes, a fixation–permeabilization protocol aimed at maximizing both preservation of protein antigenicity and accessibility of nucleic acid to DNA probes was used. Asynchronous HCT116 cells grown on coverslips were rinsed three times in PBS and fixed for 10 min with fixation buffer (0.2% Triton X-100, 20 mM Pipes pH 6.8, 1 mM MgCl2, 10 mM ethylene glycol tetraacetic acid [EGTA], 4% formaldehyde) at RT. Slides were then washed for three times 3 min with PBS containing 0.01% Triton X-100 and incubated for 5 min in 0.1 M HCl at RT. Cells were rinsed in 2× SSC and incubated in 2× SSC with 50% formamide for at least 1 h at RT before hybridization.
Immuno-FISH and immuno-fiber FISH
HCT116 cells (on coverslips) or HeLaS3 chromatin fibers (on glass slides) were brought into contact with DNA probes, and samples were sealed with coverslips and rubber cement. Thus, the following treatments denatured both probe and target DNA simultaneously. All DNA fiber preparations were denatured for 3 min, using 75°C for centromere DNA and rDNA probes and 80°C for telomeric probes. For cells, denaturation time (at the same temperatures) was increased to 5 min. Samples were subsequently incubated overnight in a wet chamber at 42°C. Then, the rubber cement was removed, and coverslips (carrying cells) or glass slides (carrying DNA fibers) were rinsed in 4× SSC/0.2% Tween and washed for three times 5 min in the same solution at 42°C, followed by three times 5 min in 1× SSC at 60°C. After rinsing in 4× SSC/0.2% Tween, coverslips were blocked for 30 min in 3% bovine serum albumin (BSA) in 4× SSC/0.2% Tween in a wet chamber at 42°C. Detection of DNA probes was performed by incubating with appropriate antibodies diluted in 3% BSA in 4× SSC/0.2% Tween for 60 min in a wet chamber at 42°C. Samples were then washed for three times 5 min in 4× SSC/0.2% Tween at 42°C and rinsed in PBS for the subsequent IF procedure. Primary antibodies were diluted in PBS and 3% BSA and added to the samples for 90 min at RT. Next, samples were washed for three times 5 min in PBS and incubated for 60 min with secondary antibodies diluted in PBS, 3% BSA. After washing for three times 5 min in PBS, samples were rinsed in 2× SSC and counterstained for 5 min with 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml).
Microscopy and image analysis
Images were captured using a Deltavision microscope on a Nikon Eclipse TE200 base (Applied Precision, Issaquah, WA) with an Plan Apo 60/1.4 oil immersion objective and a CoolSnap HQ camera (Photometrics) or on a Nikon Eclipse TE2000 equipped with the same objective and a VDS COOL-1300Q camera (Vosskuehler). Images taken at different focal planes were processed with a deconvolution algorithm and projected into one picture using the Softworx software (Applied Precision). The projected images were opened in Adobe Photoshop and then sized and placed in figures using Adobe Illustrator CS (Adobe Systems, Mountain View, CA).
ChIP assay and Southern blotting
For ChIP assays, approximately 2 × 106 HeLaS3 anaphase cells (prepared as described above) were used per experiment. Anaphase cells were first treated with 5 mM dimethyl 3,3′-dithiobispropionimidate·2 HCl/PBS for 30 min on ice, washed in PBS, and cross-linked in 1% formaldehyde/PBS at RT for 10 min. The cross-linking was then quenched by adding 10% of 1.375 M glycine to the cell suspension. After two washes with ice-cold PBS, cells were resuspended in 500 μl of SDS-lysis buffer (0.1% SDS, 0.1% sodiumdeoxycholate, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM CaCl2, 1 mM phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. Then, cells were lysed by sonication for 30 s and treated with 300 U/ml of micrococcal nuclease at 37°C for 10 min to produce chromatin fragments of less than 1 kb (<5 nucleosomes), followed by centrifugation for 15 min at 14,000 rpm. The resulting chromatin fractions (supernatants) were diluted 10× in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM ethylenediamine tetraacetic acid [EDTA], 16.7 mM Tris–HCl pH 8.0, 167 mM NaCl), and precleared with 50 μl of salmon sperm DNA-saturated protein Sepharose G beads (200 μg/ml salmon sperm DNA, 50% slurry). Then, 5-ml chromatin fractions were first incubated with 5 μg of antibody for 2 h at 4°C, followed by an overnight incubation with 100 μl of salmon sperm DNA saturated Sepharose G beads. Beads were washed twice in low salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 150 mM NaCl), twice in high salt buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.0, 500 mM NaCl), twice in LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxychlorate, 1 mM EDTA, 10 mM Tris–HCl pH 8.0), and twice in TE buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA). Bound nucleosomes were then eluted for 15 min by shaking in 1 ml elution buffer (50 mM Tris pH 8.0, 1% SDS, 1 mM EDTA, 0.1 M NaHCO3). Cross-links were reversed by adding 100 μl of 1 M dithiothreitol and incubation at 37°C for 30 min, followed by the addition of 40 μl 5 M NaCl, 20 μl 0.5 M EGTA, 40 μl 1 M Tris pH 6.5, 10 μl 10 mg/ml proteinase K, and 10 μl 10 mg/ml RNase, and incubation at 65°C for 4–6 h. DNA was recovered by phenol/chloroform extraction, precipitated by isopropanol, washed in 70% ethanol, and then dissolved in 50 μl TE buffer.
For detection of centromere DNA by Southern blotting, 10 μl of ChIP DNA was resolved on a 1% agarose gel. DNA was transferred onto Biodyne® B membrane (KPL, Gaithersburg, MD) and hybridized for 18 h at 42°C with biotinylated pan-centromere probe or rDNA probe at concentration of 50 ng/ml. The detection of biotinylated probes was done according to the manufacturer’s instructions (Detector™ AP chemiluminescent blotting kit, KPL).
Results
PICH associates preferentially with centromeric chromatin in anaphase
In recent studies, it was shown that PICH localizes to kinetochores and inner centromeres during metaphase and then to ultrathin threads during anaphase (Baumann et al. 2007; Chan et al. 2007). To extend these observations and directly demonstrate an association of PICH with centromeric chromatin during anaphase, HeLa cells were labeled with BrdU and synchronized in anaphase. Then, mechanically extended chromatin fibers were prepared from these cells and examined by IF microscopy (note that the term “fiber” is used throughout this study to refer to chromatin preparations; in contrast, the term “thread” is used to designate the PICH-positive structures seen in anaphase cells in situ; Baumann et al. 2007). Staining of extended chromatin with anti-PICH antibodies readily demonstrated colocalization of PICH with chromatin fibers, as visualized by either anti-BrdU antibodies or DAPI (Fig. 1a). Only about half of all PICH-positive fibers were BrdU positive, consistent with the fact that not all centromere DNA will be replicated in asynchronously growing cells during a 10-h labeling. More importantly, none of more than 30 BrdU/PICH-double-positive fibers showed any gaps in BrdU labeling, arguing that these fibres represented completely replicated DNA (Torres-Rosell et al. 2007).
Immuno-fiber staining showing PICH associates with centromeric chromatin fibers in anaphase. Extended chromatin fibers were prepared from either anaphase (30-min release from Noscapine, a–d, and right panel in e) or metaphase cells (5 min release from Noscapine, left panel in e) and then examined for colocalization of PICH with the indicated markers. All bars indicate 5 μm. a PICH associates with BrdU-positive anaphase DNA fibers. b A chromatin fiber showing positivity for PICH, histone H3 dimethylated at lysine 4 (2mH3K4), and CENP-A. c A PICH-positive chromatin fiber showing staining with histone H3 trimethylated at lysine 9 (3mH3K9) and CENP-A. d Chromatin fibers stained for PICH and histone H3 acetylated at lysine 18 (AcH3K18): PICH-positive thread shows no AcH3K18 staining (left) and vice versa; AcH3K18-positive fiber shows no PICH decoration (right). e INCENP associated with PICH in metaphase chromatin fibers (left) but dissociated from PICH-positive fibers in anaphase (right). f Quantification results showing the positivity of indicated markers on PICH-positive chromatin fibers
PICH-positive fibers could also be stained using antibodies against the centromere-associated histone variant CENP-A (CenH3; Amor et al. 2004; Bloom 2007). Moreover, many PICH-positive fibers were decorated with antibodies recognizing modified histone H3, di-, or trimethylated at lysines 4 (2mH3K4) and 9 (3mH3K9; Fig. 1b and c). As expected, these latter antibodies also stained PICH-negative fibers (data not shown), in agreement with the fact that di- and trimethylated H3 histones are not restricted to centromeric chromatin (Sullivan and Karpen 2004). PICH-positive fibers were not generally stained when using antibodies against histone H3 acetylated on lysine 18 (AcH3K18; Fig. 1d, left), consistent with the hypoacetylated state of centromeric chromatin (Sullivan and Karpen 2004; Baird and Farr 2006), although these antibodies readily detected acetylated histones on PICH-negative chromatin fibers (Fig. 1d, right). Finally, we compared fibers prepared from metaphase cells (5 min after release from a Noscapine block) and anaphase cells (30 min after release) for their ability to be stained by antibodies against the inner centromere protein (INCENP; Fig. 1e). This component of the so-called chromosomal passenger complex localizes to the inner centromere at metaphase but relocates to the spindle mid-zone after anaphase onset (Vagnarelli and Earnshaw 2004). Confirming the mitotic stages analyzed, INCENP was detected on more than 80% of PICH-positive fibers prepared from metaphase cells but on only 21% of PICH-positive fibers from anaphase cells (Fig. 1f). Similar quantitative analyses revealed that more than 80% of PICH-positive anaphase fibers stained simultaneously positive for histone H3, CENP-A, 2mH3K4, and 3mH3K9, whereas only 11% were positive for AcH3K18 (Fig. 1f). Taken together, these data indicate that most of the PICH-positive fibers prepared from anaphase cells represent centromeric chromatin.
To substantiate this conclusion, we next combined IF microscopy with fiber-FISH, using an alphoid satellite DNA probe (pan-centromere probe) to directly visualize centromere DNA (Dunham et al. 1992; Baird and Farr 2006). In agreement with the results shown in Fig. 1, PICH immunoreactivity colocalized with the pan-centromere probe on extended fibers prepared from anaphase cells (Fig. 2a). The same fibers could also be stained with an antibody against CENP-A, demonstrating that they comprise both alphoid satellite DNA and this centromere-specific histone H3. As expected, only a minority of all DAPI-positive chromatin fibers stained positive for PICH and the pan-centromere probe (Fig. 1a, bottom panel), confirming the specificity of both immunostaining and hybridization signals. Moreover, when using an rDNA probe for fiber-FISH, most PICH-positive fibers did not show significant hybridization (Fig. 2b), and conversely, rDNA positive fibers were generally negative for PICH (Fig. 2c). This indicates that rDNA is not a major component of PICH-positive fibers. A quantitative analysis of these results revealed costaining of more than 80% of PICH-positive fibers with the pan-centromere probe (39 of 48), compared to only 12% double positives with the rDNA probe (5 of 42). Thus, although we cannot exclude that PICH is able to associate with different DNA sequences, these data strongly argue that the majority of PICH-positive chromatin fibers comprises centromere DNA.
Immuno-fiber FISH on PICH-positive anaphase chromatin fibers and chromatin-IP with PICH antibody. All bars indicate 5 μm. a A PICH-positive anaphase fiber showing hybridization with pan-centromere probe (PanCen) and CENP-A staining. Upper panels illustrate an enlarged region from the lower panel. b, c Chromatin fibers probed with rDNA probe and stained for PICH. PICH-positive fibers show no rDNA signals (b), and rDNA-positive fibers are negative for PICH staining (c). d ChIP assay showing that PICH binds to centromeric nucleosomes. Immunoprecipitates were prepared with antibodies against the indicated proteins. Left, Southern blot using pan-centromere probe on ChIP products. Right, Southern blot using rDNA probe
To corroborate the above cytological data with biochemical evidence, we also used ChIP to visualize PICH binding to centromeric chromatin. Nucleosome-associated DNA isolated with antibodies against either histone H3, 2mH3K4, CENP-A, or PICH reacted positively with a pan-centromere probe in Southern blots, whereas control immunoprecipitates performed with antibodies against the Flag tag or the telomere-associated protein TRF2 (Smogorzewska et al. 2000) showed little or no hybridization (Fig. 2d, left). In parallel ChIP experiments, hybridization with an rDNA probe was used for the control. Whereas nucleosomes isolated with antibodies against histone H3 or 2mH3K4 showed the expected positive hybridization with the rDNA probe, the ChIP products isolated with anti-PICH, anti-CENP-A, and anti-TRF2 antibodies only yielded background hybridization (Fig. 2d, right). These ChIP data are in excellent agreement with the chromatin fiber staining and fiber-FISH data shown above and confirm that PICH associates preferentially, if not exclusively, with centromere DNA.
PICH-positive anaphase threads contain centromere DNA
The above immuno-fiber staining, immuno-fiber-FISH, and ChIP experiments concur to demonstrate that during anaphase, PICH associates preferentially with centromeric chromatin. This strongly suggests that the PICH-positive anaphase threads reported previously (Baumann et al. 2007) result from an unraveling of this PICH-positive centromeric chromatin. Because PICH-positive threads had previously been undetectable by DAPI staining (Baumann et al. 2007), we tried to improve the sensitivity of DNA detection by incubating cells with BrdU, followed by immunostaining with an anti-BrdU antibody. In these experiments, a heat treatment was included to only partially denature DNA, so that BrdU epitopes were concealed in the condensed chromatin but revealed in less compacted DNA (Chan et al. 2007). Using this procedure, PICH-positive threads could successfully be stained with anti-BrdU antibodies (Fig. 3a; see also Chan et al. 2007).
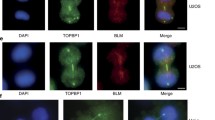
BrdU staining and FISH on anaphase cells. All bars indicate 10 μm. a Representative images showing colocalization of BrdU signals and PICH on anaphase threads. b, c HCT116 cells were subjected to FISH with pan-centromere or pan-telomere probes, followed by staining with antibodies against PICH. Merged images show PICH in green and hybridizing DNA in red. b In early anaphase, hybridization of the pan-centromere (PanCen) probe results in multiple, elongated, thread-like signals. The majority of these signals colocalize with PICH threads. This colocalization is not visible for all pan-centromere threads because of different focal planes. The lower panels show enlarged images from selected regions. c PICH-threads and pan-telomere (PanTelo) signals in early anaphase. Hybridization of a (TTAGGG) n probe results in multiple, point-like signals. In contrast to the pan-centromere probe, no elongated signals are visible, not even in early anaphase. There was no correlation between telomere signals and PICH threads at any anaphase stage
Having confirmed that PICH-positive anaphase threads comprise DNA, we next performed immunofluorescence in situ hybridization (immuno-FISH) on HCT116 cells, combining anti-PICH antibody staining with FISH hybridization using a pan-centromere probe (Fig. 3b). In cells that were judged to be in early anaphase (as inferred from the length of the PICH-positive threads), significant colocalization of PICH-positive threads and pan-centromere probe could be observed. In particular, the pan-centromere probe often produced dumbbell-like FISH signals that coincided with PICH staining (Fig. 3b). At later stages of anaphase, both the number of PICH-positive threads and the hybridization efficiency of the pan-centromere probe to the threads diminished, making it difficult to obtain satisfactory FISH signals on extended threads. We presume that this reflects a technical difficulty in performing FISH on PICH-positive threads, which apparently represent extremely decondensed, ultrathin structures that may be devoid of histones (Baumann et al. 2007; Chan et al. 2007). As a control, parallel FISH experiments were performed with a probe for telomere DNA. As shown in Fig. 3c, there was no obvious coincidence between the signals produced by the telomere probe and PICH staining. Moreover, the telomere probe did not produce the stretched and extended signals that were observed with the pan-centromere probe. Similar results were also obtained when staining HeLa cells at different stages of mitosis (Supplemental Figs. S1, S2). In these cells, the close spatial relationship between PICH-positive fibers and the signals produced by the pan-centromere but not the pan-telomere probe was also confirmed by quantitative analyses. Colocalization between PICH and pan-centromere signals was found for 95% of PICH-positive threads (255 out of 269), whereas only 7.7% of PICH threads overlapped with pan-telomere dots (21 out of 271).
Resolution of PICH-positive threads requires Topo-IIα activity specifically during anaphase
In previous experiments, PICH-positive anaphase threads were shown to be exacerbated upon inhibition of Topo-IIα in Mad2-depleted cells (Baumann et al. 2007). However, these studies did not distinguish whether Topo-IIα inhibition caused a delay in the resolution of PICH-positive threads during anaphase or, alternatively, caused the increased formation of such threads during cell cycle stages before anaphase. This distinction is critical in view of the question of when exactly during mitotic progression topoisomerase activity is required for sister chromatid separation. To address this key issue, we timed the addition of the Topo-IIα inhibitor ICRF-193 to cells carefully synchronized in anaphase and then monitored both sister chromatid separation and the resolution of PICH-positive anaphase threads. As illustrated schematically in Fig. 4a, the Topo-IIα-specific inhibitor ICRF-193 (Patel et al. 2000; Perrin et al. 1998) was added at various times during anaphase progression, before all cells were fixed for analysis. Immunostaining with antibodies against PICH and INCENP was then used to monitor the frequency of PICH-positive threads and progression through anaphase and telophase, respectively. As judged by the loss of INCENP staining from centromeres, anaphase and telophase cells accounted for more than 90% of the total mitotic population in both ICRF-193-treated and control (dimethyl sulfoxide [DMSO]-treated) cells, attesting to successful synchronization (data not shown). However, when compared to DMSO-treated control cells, the inhibition of topoisomerase activity by ICRF-193 drastically increased the frequency of PICH-positive threads and concomitantly impaired the separation of sister chromatids (Fig. 4b), confirming and extending previous results (Baumann et al. 2007). To analyze these data quantitatively, two phenotypes produced by ICRF-193 addition were distinguished. Type I cells showed numerous PICH-positive threads associated with compact chromatin masses that appeared to be either stuck in the cell center or pushed to one side of the cell; in both cases, INCENP was displaced from centromeres but not associated with a well-developed central spindle (Fig. 4b). Type II cells also showed massive PICH-positive threads, but two chromatin masses were clearly separated, and INCENP could be seen on structures resembling a central spindle or midbody, suggesting that these cells had progressed to a telophase-like stage (Fig. 4b). The frequency of these aberrations was dependent on the exact timing of ICRF-193 addition after anaphase onset. In response to early drug addition, aberrations were more frequent overall (Fig. 4c), and the more severe type I phenotype was more prevalent than the type II phenotype (Fig. 4d). These data demonstrate that topoisomerase activity is required during anaphase for both the resolution of PICH-positive threads and the complete separation of sister chromatids.
The resolution of PICH-positive anaphase threads can be inhibited by ICRF-193. a HeLa cells were first synchronized in anaphase by sequentially treatment/release from 100 mM thymidine, 20 μM noscapine, and 10 μM proteosome inhibitor MG132 at the indicated time points. DMSO or 5 μM ICRF-193 were added to cells at the indicated time points after release of cells from the MG132 block. All cells were fixed for immunostaining with PICH (red) and INCENP (green) 60 min after release from MG132. b Examples showing localizations of PICH and INCENP in control (DMSO) or ICRF-193-treated cells. Merged images show PICH in red, INCENP in green, and DAPI in blue. The dashed lines in the INCENP panels indicate the outlines of dividing cells. Two major aberrant phenotypes were identified in cells treated with ICRF-193. Type I cells showed numerous PICH-positive threads associated with chromatin that was located in the cell center (left) or pushed to one side of the cell (right). Type II cells showed a clear separation of sister chromatids, but massive PICH-positive threads remained unresolved. Bar indicates 10 μm. c Quantification of the frequency of aberrant anaphase–telophase cells (Types I + II). Bars indicate standard errors from three independent experiments. For each experiment, 250 cells were counted. d Distribution of different anaphase–telophase cells in DMSO- or ICRF-treated populations. Data were obtained from three independent experiments, counting 250 cells for each: bars denote standard errors
PICH-positive anaphase threads commonly terminate on kinetochores
To further support the proposition that most PICH-positive threads comprise centromere DNA, we carefully examined how frequently individual PICH-positive threads were connected to kinetochores during increasing chromosome separation. Anaphase cells were synchronized as described in Fig. 4a and costained with antibodies against PICH and a human kinetochore/centromere-specific CREST serum (Fritzler and Kinsella 1980; Ikeno et al. 1994). In very early anaphase cells in which the kinetochores of sister chromatids were separated by less than 2 μm, PICH localized to inner centromeres (i.e., between two dominant CREST signals) on virtually all chromosomes (Fig. 5a, left panel; Fig. 5b), as expected (Baumann et al. 2007). As cells progressed further through anaphase, the number of PICH-positive threads decreased with increasing interkinetochore distance (Fig. 5c), and concomitantly, their length increased (data not shown; see Baumann et al. 2007). Most importantly, a majority of all threads was still connected to a CREST-positive kinetochore, at least at one end (Fig. 5a, central panel; Fig. 5b). Essentially, the same conclusions were reached after analyzing ICRF-193-treated cells, except that the overall frequency of PICH-positive threads was increased at all stages of progression through anaphase and telophase (Fig. 5a, right panel; Fig. 5b and c).
PICH-positive anaphase threads often terminate at the centromere/kinetochore. a Anaphase HeLa cells were immunostained with PICH antibody and CREST antiserum. Cells were synchronized in anaphase and analyzed as described in Fig. 4a. Representative images are from samples that received DMSO or ICRF-193 20 min after release from MG132, before being analyzed 40 min later. The small images show enlarged regions (see dashed boxes). PICH-positive threads connected to CREST-positive dots were denoted as “+” while threads without CREST connection were denoted as “−”. Bar indicates 10 μm. b Quantification data showing that the majority of PICH-positive anaphase threads terminate at CREST signals, at least through one end. Interkinetochore distance was calculated by measuring the distance between separated CREST signals. White columns indicate cells treated by DMSO, and black columns show data obtained after ICRF-193 treatment. Numbers within each column indicate the total number of threads counted. c Quantification of the numbers of PICH-positive threads per cell. The numbers of cells analyzed are indicated above each column. Bars indicate standard errors. d Immunostaining of PICH and Hec1 in late anaphase cells. Top: Kinetochores connected to PICH-positive thread trail behind the bulk of the separating chromosomes; bottom: a PICH-positive thread showing a gap in the middle of the thread. Bar represents 10 μm
As expected for threads connecting sister kinetochores, the length of PICH-positive threads was positively correlated with the extent of chromosome separation (data not shown). It is interesting to note that sister kinetochores connected by PICH-positive threads in late anaphase could often be seen to trail behind the bulk of the separating chromosomes (Fig. 5d, upper panels; see also Chan et al. 2007 and Fig. 4B in Baumann et al. 2007). These observations suggest that PICH-positive threads oppose the forces exerted by the spindle microtubules, resulting in tension along the length of the thread. We also emphasize that PICH-positive threads connected most, if not all, sister kinetochores during metaphase and very early anaphase (e.g., Fig. 5a). At later stages, however, many of the threads appeared to be connected to kinetochores at only one end (data not shown), or alternatively, PICH staining showed a gap in the middle of a thread (Fig. 5d, lower panels). Although difficult to prove in the absence of live cell imaging, we interpret discontinuous threads to represent intermediates in the process of thread resolution. Specifically, we propose that decatenation of PICH-positive centromere DNA is followed by “reeling in” of this DNA, to restore a compact centromere structure on each of the separating chromosomes.
Discussion
In this paper, we have combined cytological and biochemical approaches to study, first, the association of PICH with chromatin fibers and, second, the origin and fate of ultrathin PICH-positive anaphase threads (Baumann et al. 2007; Chan et al. 2007). Our results have intriguing implications for the timing of centromere decatenation and the completion of sister chromatid separation.
What do macromolecules contribute to the formation of PICH-positive threads?
Our cytological studies on DNA fibers and ChIP experiments concur to demonstrate that PICH associates preferentially with centromeric chromatin during anaphase. Thus, in all likelihood, it is the unraveling of centromeres that leads to the formation of the ultrathin PICH-positive threads that are common in anaphase cells. In support of this conclusion, most PICH-positive threads terminate in close proximity to kinetochores, at least at the resolution of light microscopy. Although the presence of DNA within ultrathin PICH-positive threads could be demonstrated by BrdU incorporation (see also Chan et al. 2007), it has so far proven impossible to use FISH to unequivocally demonstrate that this DNA is derived from centromeres. This inability to visualize the ultrathin PICH-positive threads by FISH presumably reflects technical limitations. Hybridization to ultrathin DNA threads may result in fluorescence signals that are too weak to detect with confidence. Moreover, the need to preserve antigenicity required conditions for FISH analyses that are likely to increase background. However, we note that alphoid satellite DNA signals were reported to occasionally connect sister chromatids in salt-extracted metaphase chromosomes (Bickmore and Oghene 1996).
It is most interesting to note that in early anaphase cells, we observed strikingly elongated, stretched signals that colocalized with the PICH signal (Fig. 3b). These elongated signals were clearly distinct from the usual dot-like hybridization signals produced by pan-centromere probes on interphase cells, indicating significant conformational changes at the centromere at the onset of anaphase. The PICH signal was strongest in regions of weaker intensity of the stretched pan-centromere signal, indicating that PICH decorates the more decondensed sections (Fig. 3b). Even in late anaphase, some of the signals for the pan-centromere probe were not dot-like but stretched toward the division plane (Supplemental Fig. S1). To our knowledge, such elongated signals have not previously been reported. They were probably detected here because our preparations were highly enriched for anaphase cells, facilitating the visualization of centromere stretching at this particular cell cycle stage. Attesting to the specificity of our observation, no elongation of FISH signals was seen when a telomere probe was used for control.
With regard to the protein composition of PICH-positive threads, it is remarkable that BLM and its complex partners Topo-IIIα and hRMI1 are the only proteins that have so far been shown to associate with these structures (Chan et al. 2007). In contrast, multiple antibodies against common histones, centromere-specific histones, or histone modifications failed to decorate PICH-positive threads, raising the question of whether the DNA in these threads is associated with any histones (Baumann et al. 2007; Chan et al. 2007; Wang and Nigg, unpublished results).
What is the function of PICH-positive threads?
A priori, the unraveling of DNA to ultrathin threads during cell division is expected to pose a considerable threat to the integrity of the genome. Yet, PICH-positive threads have been seen at astonishing frequency in all cultured cell types examined so far, suggesting that they represent a physiological (rather than pathophysiological) structure. Considering the evidence presented here and elsewhere (Baumann et al. 2007; Chan et al. 2007), we indeed believe that the many short PICH-positive threads connecting sister kinetochores during early anaphase evolve directly from catenated centromeric chromatin. In contrast, the origin of some of the longest threads seen in very late anaphase is more difficult to ascertain, and it is definitely possible that some of these late persisting threads comprise incompletely replicated DNA or unresolved recombination intermediates (as discussed in Chan et al. 2007).
What advantage could cells possibly derive from the persistence of PICH-positive threads during anaphase? At present, we can propose three speculative answers, and these are not mutually exclusive. First, our data strengthen the view that persistent catenation at centromeres may cooperate with cohesin ring complexes to ensure centromere cohesion (Shamu and Murray 1992; Toyoda and Yanagida 2006). In principle, the two mechanisms may function in parallel and complement each other. This view is supported by recent data suggesting that cohesin depletion does not abolish sister centromere pairing in the absence of bidirectional pulling forces (Deehan Kenney and Heald 2006; Diaz-Martinez et al. 2007). Alternatively, cohesin proteins may function to prevent premature decatenation. Considering that Topo-IIα catalyzes both catenation and decatenation, one could argue that the removal of cohesin proteins from centromeres is required to shift the equilibrium toward decatenation. According to this scenario, the tension exerted on centromere DNA and PICH-positive threads during early anaphase would then confer directionality to the action of Topo-IIα in favor of decatenation.
Second, there is evidence that the SAC can rapidly be reactivated by addition of taxol to cells that have already completed chromosome alignment (Gorbsky and Ricketts 1993; Waters et al. 1998; Clute and Pines 1999). This suggests that maintenance of tension is important to prevent the re-establishment of an inhibitory SAC signal. Considering that PICH-positive threads oppose the pulling forces of microtubules (Fig. 5d), they contribute to maintain tension and may thereby ensure the continued silencing of the SAC.
Third, PICH-positive threads may conceivably relate to the recently proposed NoCut pathway identified in budding yeast (Norden et al. 2006). This pathway was proposed to monitor the positioning of chromatin to prevent premature ingression of the cleavage furrow. If a similar pathway operates in vertebrates, PICH-positive threads might serve as a signaling platform for the operation of this proposed checkpoint. However, we have so far been unable to detect Aurora B on PICH-positive threads, although this kinase was reported to play a key role in the NoCut pathway (Norden et al. 2006).
Finally, we note that the persistence of catenated centromeres at anaphase onset has obvious implications for the putative “decatenation checkpoint” that has been proposed to operate during G2 to prevent cells from entering mitosis with entangled DNA (Clarke et al. 2006; Damelin and Bestor 2007). Whatever structural feature this putative checkpoint monitors, our data would argue that it must be insensitive to the persistence of catenated centromeres.
How are PICH-positive threads resolved?
We cannot exclude that some PICH-positive threads are occasionally cut by nucleases or ruptured by mechanical forces. However, this would almost inevitably result in the loss of centromere-related DNA. Consequently, centromeres would shorten over successive cell generations, reminiscent of telomeres. Although centromere shortening is a very intriguing possibility, we are not presently aware of evidence that would support its occurrence. In any case, the resolution of PICH-positive threads through cuts or breakage would be expected to activate a DNA damage checkpoint, followed by double-strand break repair. It is not impossible that some instances of delayed cytokinesis might be explained by checkpoint activation in response to the breakage of PICH-positive threads.
According to our data, decatenation represents the predominant mechanism for resolving PICH-positive threads under physiological conditions. This may involve either Topo-IIα (our present study; see also Baumann et al. 2007) or Topo-IIIα, in a complex with BLM and RMI1 (Chan et al. 2007). Subsequently, we envision that the decatenated DNA is “reeled in” to join the bulk of the compact centromere on each sister chromatid. To ensure the rapid, accurate, and efficient execution of this process, it is possible that it involves motor proteins (including perhaps PICH itself?) and central spindle microtubules to provide mechanical support.
Clearly, the discovery of PICH-positive threads in anaphase cells has raised many intriguing questions that we are only beginning to answer. At present, PICH constitutes the best marker for visualizing ultrathin anaphase threads, although it is possible that PICH-positive threads correspond to structures that have occasionally been visualized in earlier cytological studies (e.g., Roos 1973; Rattner et al. 1988; Suja et al. 1992). It is interesting to note that the BLM protein also associates with PICH-positive threads at later stages of mitosis, but PICH-positive threads are also present (and in fact exacerbated) in BLM-deficient cells, indicating that the BLM complex is not an essential component of ultrathin anaphase threads (Chan et al. 2007). Major challenges for the future concern the identification of additional proteins on PICH-positive threads, which may help illuminate both the origins and the functions of these threads. In parallel, our findings invite renewed attention on the role of DNA catenation in centromere cohesion and SAC signaling, as well as the regulation of topoisomerases during mitotic progression.
References
Amor DJ, Kalitsis P, Sumer H, Choo KH (2004) Building the centromere: from foundation proteins to 3D organization. Trends Cell Biol 14(7):359–368
Baird DM, Farr CJ (2006) The organization and function of chromosomes. EMBO Rep 7(4):372–376
Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128(1):101–114
Bickmore WA, Oghene K (1996) Visualizing the spatial relationships between defined DNA sequences and the axial region of extracted metaphase chromosomes. Cell 84(1):95–104
Bloom K (2007) Centromere dynamics. Curr Opin Genet Dev 17(2):151–156
Bloom K, Sharma S, Dokholyan NV (2006) The path of DNA in the kinetochore. Curr Biol 16(8):R276–R278
Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15(11):589–598
Chan KL, North PS, Hickson ID (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. Embo J 26(14):3397–3409
Clarke DJ, Johnson RT, Downes CS (1993) Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J Cell Sci 105(Pt 2):563–569
Clarke DJ, Vas AC, Andrews CA, Diaz-Martinez LA, Gimenez-Abian JF (2006) Topoisomerase II checkpoints: universal mechanisms that regulate mitosis. Cell Cycle 5(17):1925–1928
Clute P, Pines J (1999) Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol 1(2):82–87
Damelin M, Bestor TH (2007) The decatenation checkpoint. Br J Cancer 96(2):201–205
Deehan Kenney R, Heald R (2006) Essential roles for cohesin in kinetochore and spindle function in Xenopus egg extracts. J Cell Sci 119(Pt 24):5057–5066
Diaz-Martinez LA, Gimenez-Abian JF, Clarke DJ (2007) Cohesin is dispensable for centromere cohesion in human cells. PLoS ONE 2:e318
Dunham I, Lengauer C, Cremer T, Featherstone T (1992) Rapid generation of chromosome-specific alphoid DNA probes using the polymerase chain reaction. Hum Genet 88(4):457–462
Fritzler MJ, Kinsella TD (1980) The CREST syndrome: a distinct serologic entity with anticentromere antibodies. Am J Med 69(4):520–526
Gandhi R, Gillespie PJ, Hirano T (2006) Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol 16(24):2406–2417
Gorbsky GJ, Ricketts WA (1993) Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J Cell Biol 122(6):1311–1321
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19(17):4780
Ikeno M, Masumoto H, Okazaki T (1994) Distribution of CENP-B boxes reflected in CREST centromere antigenic sites on long-range alpha-satellite DNA arrays of human chromosome 21. Hum Mol Genet 3(8):1245–1257
Kitajima TS, Sakuno T, Ishiguro K, Iemura S, Natsume T et al (2006) Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature 441(7089):46–52
Kops GJ, Weaver BA, Cleveland DW (2005) On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer 5(10):773–785
Koshland D, Hartwell LH (1987) The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science 238(4834):1713–1716
Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A et al (2006) Wapl controls the dynamic association of cohesin with chromatin. Cell 127(5):955–967
Langer S, Fauth C, Rocchi M, Murken J, Speicher MR (2001) AcroM fluorescent in situ hybridization analyses of marker chromosomes. Hum Genet 109(2):152–158
Losada A (2007) Cohesin regulation: fashionable ways to wear a ring. Chromosoma 116(4):321–329
Losada A, Hirano M, Hirano T (2002) Cohesin release is required for sister chromatid resolution, but not for condensin-mediated compaction, at the onset of mitosis. Genes Dev 16(23):3004–3016
Murray AW, Szostak JW (1985) Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol 1:289–315
Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8(5):379–393
Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74:595–648
Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S et al (2006) The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell 125(1):85–98
Patel S, Jazrawi E, Creighton AM, Austin CA, Fisher LM (2000) Probing the interaction of the cytotoxic bisdioxopiperazine ICRF-193 with the closed enzyme clamp of human topoisomerase IIalpha. Mol Pharmacol 58(3):560–568
Perrin D, van Hille B, Hill BT (1998) Differential sensitivities of recombinant human topoisomerase IIalpha and beta to various classes of topoisomerase II-interacting agents. Biochem Pharmacol 56(4):503–507
Rattner JB, Kingwell BG, Fritzler MJ (1988) Detection of distinct structural domains within the primary constriction using autoantibodies. Chromosoma 96(5):360–367
Riedel CG, Katis VL, Katou Y, Mori S, Itoh T et al (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature 441(7089):53–61
Roos UP (1973) Light and electron microscopy of rat kangaroo cells in mitosis. I. Formation and breakdown of the mitotic apparatus. Chromosoma 40(1):43–82
Seki M, Nakagawa T, Seki T, Kato G, Tada S et al (2006) Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol 26(16):6299–6307
Shamu CE, Murray AW (1992) Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol 117(5):921–934
Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR et al (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20(5):1659–1668
Suja JA, Antonio C, Rufas JS (1992) Involvement of chromatid cohesiveness at the centromere and chromosome arms in meiotic chromosome segregation: a cytological approach. Chromosoma 101(8):493–501
Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11(11):1076–1083
Sumara I, Vorlaufer E, Stukenberg PT, Kelm O, Redemann N et al (2002) The dissociation of cohesin from chromosomes in prophase is regulated by Polo-like kinase. Mol Cell 9(3):515–525
Sundin O, Varshavsky A (1981) Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell 25(3):659–669
Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC et al (2006) PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell 10(5):575–585
Torres-Rosell J, De Piccoli G, Cordon-Preciado V, Farmer S, Jarmuz A et al (2007) Anaphase onset before complete DNA replication with intact checkpoint responses. Science 315(5817):1411–1415
Toyoda Y, Yanagida M (2006) Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol Biol Cell 17(5):2287–2302
Vagnarelli P, Earnshaw WC (2004) Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113(5):211–222
Waters JC, Chen RH, Murray AW, Salmon ED (1998) Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol 141(5):1181–1191
Wu L, Hickson ID (2003) The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426(6968):870–874
Wu L, Bachrati CZ, Ou J, Xu C, Yin J et al (2006) BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA 103(11):4068–4073
Yanagida M (2005) Basic mechanism of eukaryotic chromosome segregation. Philos Trans R Soc Lond B Biol Sci 360(1455):609–621
Acknowledgments
We thank Christoph Baumann for his participation at the early stage of this project and Sabine Elowe, Manuel Kaulich, and Ulf Klein for helpful discussions. This work was supported by the Max-Planck Society and the “Fonds der Chemischen Industrie.” Lily H-C W. acknowledges a fellowship from the Taiwan Merit Scholarships program (NSC-095-SAF-I-564–627-TMS).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Communicated by A. Choo
Wang and Schwarzbraun both contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. S1
PICH vs pan-centromere probe in HeLa cells. HeLa cells at different cell cycle stages were subjected to FISH with a pan-centromere probe, followed by staining with antibodies against PICH. Merged images show PICH in green, hybridizing DNA in red, and bulk DNA (DAPI staining) in blue. The interphase panel shows projected images from a Z-stack through a whole cell, whereas mitotic panels show single focal planes. Bar indicates 10 μm (GIF 164 KB)
Supplemental Fig. S2
PICH vs pan-telomeric probe in HeLa cells. All panels show projected images from Z-stacks. Bar denotes 10 μm. Note that unlike the pan-centromere probe, the pan-telomere probe yielded dot-like rather than elongated, thread-like hybridization signals (GIF 179 KB)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Wang, L.HC., Schwarzbraun, T., Speicher, M.R. et al. Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma 117, 123–135 (2008). https://doi.org/10.1007/s00412-007-0131-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-007-0131-7