Abstract
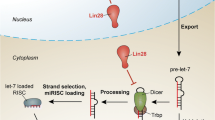
MicroRNAs (miRNAs) were first discovered in genetic screens for regulators of developmental timing in the stem-cell-like seam cell lineage in Caenorhabditis elegans. As members of the heterochronic pathway, the lin-4 and let-7 miRNAs are required in the seam cells for the correct progression of stage-specific events and to ensure that cell cycle exit and terminal differentiation occur at the correct time. Other heterochronic genes such as lin-28 and lin-41 are direct targets of the lin-4 and let-7 miRNAs. Recent findings on the functions of the let-7 and lin-4/mir-125 miRNA families and lin-28 and lin-41 orthologs from a variety of organisms suggest that core elements of the heterochronic pathway are retained in mammalian stem cells and development. In particular, these genes appear to form bistable switches via double-negative feedback loops in both nematode and mammalian stem cell development, the functional relevance of which is finally becoming clear. let-7 inhibits stem cell self-renewal in both normal and cancer stem cells of the breast and acts as a tumor suppressor in lung and breast cancer. let-7 also promotes terminal differentiation at the larval to adult transition in both nematode stem cells and fly wing imaginal discs and inhibits proliferation of human lung and liver cancer cells. Conversely, LIN-28 is a highly specific embryonic stem cell marker and is one of four “stemness” factors used to reprogram adult fibroblasts into induced pluripotent stem cells; furthermore, lin-28 is oncogenic in hepatocellular carcinomas. Therefore, a core module of heterochronic genes—lin-28, lin-41, let-7, and lin-4/mir-125—acts as an ancient regulatory switch for differentiation in stem cells (and in some cancers), illustrating that nematode seam cells mirror miRNA regulatory networks in mammalian stem cells during both normal development and cancer.




Similar content being viewed by others
References
Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell 9:403–414
Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell 4:625–637
Ambros V (2001) microRNAs: tiny regulators with great potential. Cell 107:823–826
Ambros V, Horvitz HR (1984) Heterochronic mutants of the nematode Caenorhabditis elegans. Science (New York, NY) 226:409–416
Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13:807–818
Balzer E, Moss EG (2007) Localization of the developmental timing regulator Lin28 to mRNP complexes, P-bodies and stress granules. RNA Biol 4:16–25
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Gen 40:499–507
Betschinger J, Mechtler K, Knoblich JA (2006) Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 124:1241–1253
Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ (2007) Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 101:2999–3004
Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18:943–950
Chang TC, Zeitels LR, Hwang HW, Chivukula RR, Wentzel EA, Dews M, Jung J, Gao P, Dang CV, Beer MA et al (2009) Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc Natl Acad Sci USA 106:3384–3389
Chin LJ, Ratner E, Leng S, Zhai R, Nallur S, Babar I, Muller RU, Straka E, Su L, Burki EA et al (2008) A SNP in a let-7 microRNA complementary site in the KRAS 3′ untranslated region increases non-small cell lung cancer risk. Cancer Res 68:8535–8540
Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R et al (2008) An endogenous small interfering RNA pathway in Drosophila. Nature 453:798–802
Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G et al (2006) Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell 124:343–354
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev 6:259–269
Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ (2005) Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn 234:868–877
Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ (2008) The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle (Georgetown, Tex 7:759–764
Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z et al (2008) Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science (New York, NY) 320:1077–1081
Girard A, Sachidanandam R, Hannon GJ, Carmell MA (2006) A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202
Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23–34
Grivna ST, Beyret E, Wang Z, Lin H (2006) A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20:1709–1714
Grosshans H, Johnson T, Reinert KL, Gerstein M, Slack FJ (2005) The temporal patterning microRNA let-7 regulates several transcription factors at the larval to adult transition in C. elegans. Dev Cell 8:321–330
Guo Y, Chen Y, Ito H, Watanabe A, Ge X, Kodama T, Aburatani H (2006) Identification and characterization of lin-28 homolog B (LIN28B) in human hepatocellular carcinoma. Gene 384:51–61
Hallam SJ, Jin Y (1998) lin-14 regulates the timing of synaptic remodelling in Caenorhabditis elegans. Nature 395:78–82
Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V (2009) nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell 136:926–938
Hayes GD, Ruvkun G (2006) Misexpression of the Caenorhabditis elegans miRNA let-7 is sufficient to drive developmental programs. Cold Spring Harbor Symp Quant Biol 71:21–27
Heo I, Joo C, Cho J, Ha M, Han J, Kim VN (2008) Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol Cell 32:276–284
Ibarra I, Erlich Y, Muthuswamy SK, Sachidanandam R, Hannon GJ (2007) A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev 21:3238–3243
Jeon M, Gardner HF, Miller EA, Deshler J, Rougvie AE (1999) Similarity of the C. elegans developmental timing protein LIN-42 to circadian rhythm proteins. Science (New York, NY) 286:1141–1146
Jin Z, Xie T (2007) Dcr-1 maintains Drosophila ovarian stem cells. Curr Biol 17:539–544
Johnson SM, Lin SY, Slack FJ (2003) The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol 259:364–379
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ (2005) RAS is regulated by the let-7 microRNA family. Cell 120:635–647
Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J et al (2007) The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67:7713–7722
Kanamoto T, Terada K, Yoshikawa H, Furukawa T (2006) Cloning and regulation of the vertebrate homologue of lin-41 that functions as a heterochronic gene in Caenorhabditis elegans. Dev Dyn 235:1142–1149
Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K (2005) Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19:489–501
Kim VN, Han J, Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10:126–139
Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH (2004) Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res 32:6284–6291
Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH (2006) In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods 3:27–29
Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. (2006) Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 442:818–822
Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA 105:3903–3908
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science (New York, NY) 294:853–858
Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T (2002) Identification of tissue-specific microRNAs from mouse. Curr Biol 12:735–739
Lancman JJ, Caruccio NC, Harfe BD, Pasquinelli AE, Schageman JJ, Pertsemlidis A, Fallon JF (2005) Analysis of the regulation of lin-41 during chick and mouse limb development. Dev Dyn 234:948–960
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science (New York, NY) 294:858–862
Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE (2006) Characterization of the piRNA complex from rat testes. Science (New York, NY) 313:363–367
Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science (New York, NY) 294:862–864
Lee YS, Dutta A (2007) The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 21:1025–1030
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75:843–854
Lee YS, Kim HK, Chung S, Kim KS, Dutta A (2005) Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280:16635–16641
Leung AK, Sharp PA (2006) Function and localization of microRNAs in mammalian cells. Cold Spring Harbor Symp Quant Biol 71:29–38
Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ (2003) The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell 4:639–650
Lin YC, Hsieh LC, Kuo MW, Yu J, Kuo HH, Lo WL, Lin RJ, Yu AL, Li WH (2007) Human TRIM71 and its nematode homologue are targets of let-7 microRNA and its zebrafish orthologue is essential for development. Mol Biol Evol 24:2525–2534
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435:834–838
Maller Schulman BR, Liang X, Stahlhut C, DelConte C, Stefani G, Slack FJ (2008) The let-7 microRNA target gene, Mlin41/Trim71 is required for mouse embryonic survival and neural tube closure. Cell Cycle (Georgetown, Tex 7:3935–3942
Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G et al (2004) MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet 36:1079–1083
Mayr C, Hemann MT, Bartel DP (2007) Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science (New York, NY) 315:1576–1579
Meroni G, Diez-Roux G (2005) TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147–1157
Morita K, Han M (2006) Multiple mechanisms are involved in regulating the expression of the developmental timing regulator lin-28 in Caenorhabditis elegans. EMBO J 25:5794–5804
Moss EG (2007) Heterochronic genes and the nature of developmental time. Curr Biol 17:R425–R434
Moss EG, Lee RC, Ambros V (1997) The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell 88:637–646
Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA (2008) Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature 454:241–245
Newman MA, Thomson JM, Hammond SM (2008) Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA (New York, NY) 14:1539–1549
O'Farrell F, Kylsten P (2008) A mis-expression study of factors affecting Drosophila PNS cell identity. Biochem Biophys Res Commun 370:657–662
O'Farrell F, Esfahani SS, Engstrom Y, Kylsten P (2008) Regulation of the Drosophila lin-41 homologue dappled by let-7 reveals conservation of a regulatory mechanism within the LIN-41 subclade. Dev Dyn 237:196–208
Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC (2008) The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453:803–806
Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, Dinulescu DM, Lengyel E, Peter ME (2007) Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle (Georgetown, Tex) 6:2585–2590
Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P et al (2000) Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 408:86–89
Pepper AS, McCane JE, Kemper K, Yeung DA, Lee RC, Ambros V, Moss EG (2004) The C. elegans heterochronic gene lin-46 affects developmental timing at two larval stages and encodes a relative of the scaffolding protein gephyrin. Development 131:2049–2059
Piskounova E, Viswanathan SR, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gregory RI (2008) Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J Biol Chem 283:21310–21314
Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A (2007) Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev 21:1125–1138
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403:901–906
Richards M, Tan SP, Tan JH, Chan WK, Bongso A (2004) The transcriptome profile of human embryonic stem cells as defined by SAGE. Stem Cells (Dayton, Ohio) 22:51–64
Rougvie AE (2005) Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development 132:3787–3798
Roush S, Slack FJ (2008) The let-7 family of microRNAs. Trends Cell Biol 18:505–516
Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP (2006) Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127:1193–1207
Ruvkun G (2008) The perfect storm of tiny RNAs. Nat Med 14:1041–1045
Ruvkun G, Wightman B, Ha I (2004) The 20 years it took to recognize the importance of tiny RNAs. Cell 116:S93–S96 92 p following S96
Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG (2008) A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol 10:987–993
Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science (New York, NY) 320:1643–1647
Schulman BR, Esquela-Kerscher A, Slack FJ (2005) Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn 234:1046–1054
Schwamborn JC, Berezikov E, Knoblich JA (2009) The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell 136:913–925
Seggerson K, Tang L, Moss EG (2002) Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol 243:215–225
Sempere LF, Dubrovsky EB, Dubrovskaya VA, Berger EM, Ambros V (2002) The expression of the let-7 small regulatory RNA is controlled by ecdysone during metamorphosis in Drosophila melanogaster. Dev Biol 244:170–179
Sempere LF, Sokol NS, Dubrovsky EB, Berger EM, Ambros V (2003) Temporal regulation of microRNA expression in Drosophila melanogaster mediated by hormonal signals and broad-Complex gene activity. Dev Biol 259:9–18
Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biology 5:R13
Sharp PA (2001) RNA interference—2001. Genes Dev 15:485–490
Slack F, Ruvkun G (1997) Temporal pattern formation by heterochronic genes. Annu Rev Genet 31:611–634
Slack FJ, Weidhaas JB (2006) MicroRNAs as a potential magic bullet in cancer. Future Oncol (London, England) 2:73–82
Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G (2000) The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell 5:659–669
Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG (2005) Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21:1469–1477
Sokol NS, Xu P, Jan YN, Ambros V (2008) Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 22:1591–1596
Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9:219–230
Sulston JE, Horvitz HR (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev Biol 56:110–156
Sulston JE, Albertson DG, Thomson JN (1980) The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev Biol 78:542–576
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y et al (2004) Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res 64:3753–3756
Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM et al (2008) Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453:534–538
Thomson JM, Parker J, Perou CM, Hammond SM (2004) A custom microarray platform for analysis of microRNA gene expression. Nat Methods 1:47–53
Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20:2202–2207
Vasudevan S, Steitz JA (2007) AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128:1105–1118
Vasudevan S, Tong Y, Steitz JA (2007) Switching from repression to activation: microRNAs can up-regulate translation. Science (New York, NY) 318:1931–1934
Vasudevan S, Tong Y, Steitz JA (2008) Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle (Georgetown, Tex) 7:1545–1549
Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004a) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′ UTR. Genes Dev 18:132–137
Vella MC, Reinert K, Slack FJ (2004b) Architecture of a validated microRNA::target interaction. Chem Biol 11:1619–1623
Visvader JE, Lindeman GJ (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev 8:755–768
Viswanathan SR, Daley GQ, Gregory RI (2008) Selective blockade of microRNA processing by Lin28. Science (New York, NY) 320:97–100
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39:380–385
Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH (2005) MicroRNA expression in zebrafish embryonic development. Science (New York, NY) 309:310–311
Wu L, Belasco JG (2005) Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol Cell Biol 25:9198–9208
Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R (2007) Post-transcriptional regulation of the let-7 microRNA during neural cell specification. Faseb J 21:415–426
Xu B, Zhang K, Huang Y (2009) Lin28 modulates cell growth and associates with a subset of cell cycle regulator mRNAs in mouse embryonic stem cells. RNA (New York, NY) 15:357–361
Yang DH, Moss EG (2003) Temporally regulated expression of Lin-28 in diverse tissues of the developing mouse. Gene Expr Patterns 3:719–726
Yokoyama S, Hashimoto M, Shimizu H, Ueno-Kudoh H, Uchibe K, Kimura I, Asahara H (2008) Dynamic gene expression of Lin-28 during embryonic development in mouse and chicken. Gene Expr Patterns 8:155–160
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J et al (2007a) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131:1109–1123
Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R et al (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science (New York, NY) 318:1917–1920
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Rights and permissions
About this article
Cite this article
Nimmo, R.A., Slack, F.J. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma 118, 405–418 (2009). https://doi.org/10.1007/s00412-009-0210-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0210-z