Abstract
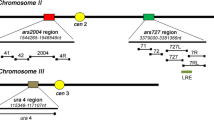
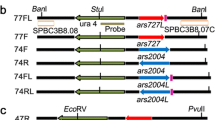
We have previously peported that the replication orgin region located near the ura4 gene on chromosome III of the fission yeast, Schizosaccharomyces pombe, contains three closely spaced origins, each associated with an autonomously replicating sequence (ARS) element. Here we report the nucleotide sequences of two of these ARS elements, ars3002 and ars3003. The two ARS elements are located on either side of a transcribed 1.5 kb open reading frame. Like 11 other S. pombe ARS elements whose sequences have previously been determined in other laboratories, the 2 new ARS elements are unusually A+T-rich. All 13 ARS elements contain easily unwound stretches of DNA. Each of the ARS elements contains numerous copies, at a higher than expected frequency, of short stretches of A+T-rich DNA in which most of the Ts are on one strand and most of the As are on the complementary strand. We discuss the potential significance for ARS function of these multiple asymmetric A+T-rich sequences.
Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amati B, Gasser SM (1990) Drosophila scaffold-attached regions bind nuclear scaffolds and can function as ARS elements in both budding and fission yeasts. Mol Cell Biol 10:5442–5454
Amati BB, Gasser SM (1988) Chromosomal ARS and CEN elements bind specifically to the yeast nuclear scaffold. Cell 54:967–978
Barker DG, White JHM, Johnston LH (1987) Molecular characterisation of the DNA ligase gene, CDC17, from the fission yeast Schizosaccharomyces pombe. Eur J Biochem 162:659–667
Bell SP, Stillman B (1992) ATP dependent recognition of eukaryotic orgins of DNA replication by a multi-protein complex. Nature 357:128–134
Breslauer KJ, Frank R, Blöcker H, Marky LA (1986) Predicting DNA duplex stability from the base sequence. Proc Natl Acad Sci USA 83:3746–3750
Brewer BJ, Fangman WL (1987) The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463–471
Campbell JL, Newlon CS (1991) Chromosomal DNA replication. In: Broach JR, Pringle JR, Jones EW (eds) The molecular biology and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. (Cold Spring Harbor monograph series 1) Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 41–146
Deshpande AM, Newlon CS (1992) The ARS consensus sequence is required for chromosomal origin function in Saccharomyces cerevisiae. Mol Cell Biol 12:4305–4313
Dubey DD, Zhu J, Carlson DL, Sharma K, Huberman JA (1994) Three ARS elements contribute to the ura4 replication origin region in the fission yeast, Schizosaccharomyces pombe. EMBO J 13:3638–3647
Eckdahl TT, Anderson JN (1990) Conserved DNA structures in origins of replication. Nucleic Acids Res 18:1609–1612
Gasser SM, Laemmli UK (1986) Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell 46:521–530
Grimm C, Kohli J (1988) Observations on integrative transformation in Schizosaccharomyces pombe. Mol Gen Genet 215:87–93
Grimm C, Kohli J, Murray J, Maundrell K (1988) Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol Gen Genet 215:81–86
Heyer W-D, Sipiczki M, Kohli J (1986) Replicating plasmids in Schizosaccharomyces pombe: Improvement of symmetric segregation by a new genetic element. Mol Cell Biol 6:80–89
Hoffman CS, Winston F (1987) A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267–272
Huang R-Y, Kowalski D (1993) A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J 12:4521–4531
Huberman JA, Spotila LD, Nawotka KA, El-Assouli SM, Davis LR (1987) The in vivo replication origin of the yeast 2 μm plasmid. Cell 51:473–481
Jacob F, Brenner S, Cuzin F (1963) On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol 28:329–348
Johnston LH, Barker DG (1987) Characterisation of an autonomously replicating sequence from the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 207:161–164
Luehrsen KR, Pearlman RE, Pata J, Orias E (1988) Comparison of Tetrahymena ARS sequence function in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe. Curr Genet 14:225–233
Marahrens Y, Stillman B (1992) A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255:817–823
Maundrell K, Wright APH, Piper M, Shall S (1985) Evaluation of heterologous ARS activity in S. cerevisiae using cloned DNA from S. pombe. Nucleic Acids Res 13:3711–3722
Maundrell K, Hutchison A, Shall S (1988) Sequence analysis of ARS elements in fission yeast. EMBO J 7:2203–2209
Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast. Methods Enzymol 194:795–823
Natale DA, Schubert AE, Kowalski D (1992) DNA helical stability accounts for mutational defects in a yeast replication origin. Proc Natl Acad Sci USA 89:2654–2658
Natale DA, Umek RM, Kowalski D (1993) Ease of DNA unwinding is a conserved property of yeast replication origins. Nucleic Acids Res 21:555–560
Nelson HCM, Finch JT, Luisi BF, Klug A (1987) The structure of an oligo(dA)-oligo(dT) tract and its biological implications. Nature 330:221–226
Pardoll DM, Vogelstein B, Coffey DS (1980) A fixed site of DNA replication in eucaryotic cells. Cell 19:527–536
Pearson WR, Lipman DJ (1988) Improved tools for biological sequence comparison. Proc Natl Acad Sci USA 85:2444–2448
Razin SV, Kekelidze MG, Lukanidin EM, Scherrer K, Georgiev GP (1986) Replication origins are attached to the nuclear skeleton. Nucleic Acids Res 14:8189–8207
Rivier DH, Rine J (1992) An origin of DNA replication and a transcription silencer require a common element. Science 256:659–663
Saito Y, Laemmli UK (1994) Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell 76:609–622
Sakaguchi J, Yamamoto M (1982) Cloned ura1 locus of Schizosaccharomyces pombe propagates autonomously in this yeast assuming a polymeric form. Proc Natl Acad Sci USA 79:7819–7823
Sakai K, Sakaguchi J, Yamamoto M (1984) High-frequency co-transformation by copolymerization of plasmids in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 4:651–656
Snyder M, Buchman AR, Davis RW (1986) Bent DNA at a yeast autonomously replicating sequence. Nature 324:87–89
Umek RM, Kowalski D (1988) The ease of DNA unwinding as a determinant of initiation at yeast replication origins. Cell 52:559–567
Umek RM, Kowalski D (1990) Thermal energy suppresses mutational defects in DNA unwinding at a yeast replication origin. Proc Natl Acad Sci USA 87:2486–2490
Walker SS, Malik AK, Eisenberg S (1991) Analysis of the interactions of functional domains of a nuclear origin of replication from Saccharomyces cerevisiae. Nucleic Acids Res 19:6255–6262
Wright APH, Maundrell K, Shall S (1986) Transformation of Schizosaccharomyces pombe by non-homologous, unstable integration of plasmids in the genome. Curr Genet 10:503–508
Zhu J, Brun C, Kurooka H, Yanagida M, Huberman JA (1992) Identification and characterization of a complex chromosomal replication origin in Schizosaccharomyces pombe. Chromosoma 102:S7-S16
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Zhu, J., Carlson, D.L., Dubey, D.D. et al. Comparison of the two major ARS elements of the ura4 replication origin region with other ARS elements in the fission yeast, Schizosaccharomyces pombe . Chromosoma 103, 414–422 (1994). https://doi.org/10.1007/BF00362286
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00362286