Abstract
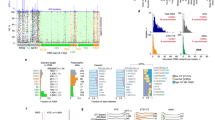
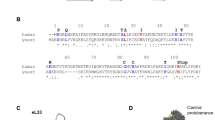
5S rRNA extends from the central protuberance of the large ribosomal subunit, through the A-site finger, and down to the GTPase-associated center. Here, we present a structure-function analysis of seven 5S rRNA alleles which are sufficient for viability in the yeast Saccharomyces cerevisiae when expressed in the absence of wild-type 5S rRNAs, and extend this analysis using a large bank of mutant alleles that show semi-dominant phenotypes in the presence of wild-type 5S rRNA. This analysis supports the hypothesis that 5S rRNA serves to link together several different functional centers of the ribosome. Data are also presented which suggest that in eukaryotic genomes selection has favored the maintenance of multiple alleles of 5S rRNA, and that these may provide cells with a mechanism to post-transcriptionally regulate gene expression.









Similar content being viewed by others
References
Altschul SF, Gish W, Miller E, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4Å resolution. Science 289:905–920
Betzel C, Lorenz S, Furste JP, Bald R, Zhang M, Schneider TR, Wilson KS, Erdmann VA (1994) Crystal structure of domain A of Thermus flavus 5S rRNA and the contribution of water molecules to its structure. FEBS Lett 351:159–164
Boeke JD, Xu H, Fink GR (1988) A general method for the chromosomal amplification of genes into yeast. Science 239:280–282
Bogdanov AA, Dontsova OA, Dokudovskaya SS, Lavrik IN (1995) Structure and function of 5S rRNA in the ribosome. Biochem Cell Biol 73:869–876
Brow DA, Geiduschek EP (1987) Modulation of yeast 5S rRNA synthesis in vitro by ribosomal protein YL3. J Biol Chem 262:13953–13958
Carroll K, Wickner RB (1995) Translation and M1 dsRNA propagation: MAK18 = RPL41B and cycloheximide curing. J Bacteriol 177:2887–2891
Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P (1992) Multifunctional yeast high-copy-number shuttle vectors. Yeast 110:119–122
Deshmukh M, Tsay YF, Paulovich AG, Woolford JL, Jr (1993) Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol Cell Biol 13:2835–2845
Deshmukh M, Stark J, Yeh LC, Lee JC, Woolford JL Jr (1995) Multiple regions of yeast ribosomal protein L1 are important for its interaction with 5S rRNA and assembly into ribosomes. J Biol Chem 270:30148–30156
Dinman JD (1995) Ribosomal frameshifting in yeast viruses. Yeast 11:1115–1127
Dinman JD, Wickner RB (1992) Ribosomal frameshifting efficiency and Gag/Gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol 66:3669–3676
Dinman JD, Wickner RB (1995) 5S rRNA is involved in fidelity of translational reading frame. Genetics 141:95–105
Dinman JD, Icho T, Wickner RB (1991) A −1 ribosomal frameshift in a double-stranded RNA virus forms a Gag-pol fusion protein. Proc Natl Acad Sci USA 88:174–178
Dinman JD, Ruiz-Echevarria MJ, Czaplinski K, Peltz SW (1997) Peptidyl transferase inhibitors have antiviral properties by altering programmed −1 ribosomal frameshifting efficiencies: development of model systems. Proc Natl Acad Sci USA 94:6606–6611
Dokudovskaya S, Dontsova O, Shpanchenko O, Bogdanov A, Brimacombe R (1996) Loop IV of 5S ribosomal RNA has contacts both to domain II and to domain V of the 23S RNA. RNA 2:146–152
Farabaugh PJ (1996) Programmed translational frameshifting. Microbiol Rev 60:103–134
Ford PJ, Southern EM (1973) Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol 241:7–12
Frank J (2003) Electron microscopy of functional ribosome complexes. Biopolymers 68:223–233
Funari SS, Rapp G, Perbandt M, Dierks K, Vallazza M, Betzel C, Erdmann VA, Svergun DI (2000) Structure of free Thermus flavus 5S rRNA at 1.3 nm resolution from synchrotron X-ray solution scattering. J Biol Chem 275:31283–31288
Harger JW, Dinman JD (2003) An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9:1019–1024
Harger JW, Dinman JD (2004) Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA 10:1721–1729
Harger JW, Meskauskas A, Dinman JD (2002) An ’integrated model’ of programmed ribosomal frameshifting and post-transcriptional surveillance. Trends Biochem Sci 27:448–454
Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A (2001) High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107:679–688
Huber PW, Rife JP, Moore PB (2001) The structure of helix III in Xenopus oocyte 5S rRNA: an RNA stem containing a two-nucleotide bulge. J Mol Biol 312:823–832
Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Jacobs JL, Dinman JD (2004) Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res 32:e160–e170
Kawakami K, Paned S, Faioa B, Moore DP, Boeke JD, Farabaugh PJ, Strathern JN, Nakamura Y, Garfinkel DJ (1993) A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135:309–320
Kinzy TG, Harger JW, Carr-Schmid A, Kwon J, Shastry M, Justice MC, Dinman JD (2002) New targets for antivirals: the ribosomal A-site and the factors that interact with it. Virology 300:60–70
Lorenz S, Perbandt M, Lippmann C, Moore K, DeLucas LJ, Betzel C, Erdmann VA (2000) Crystallization of engineered Thermus flavus 5S rRNA under earth and microgravity conditions. Acta Crystallogr D Biol Crystallogr 56:498–500
Meskauskas A, Dinman JD (2001) Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7:1084–1096
Meskauskas A, Baxter JL, Carr EA, Yasenchak J, Gallagher JEG, Baserga SJ, Dinman JD (2003a) Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol Cell Biol 23:1602–1613
Meskauskas A, Harger JW, Jacobs KLM, Dinman JD (2003b) Decreased peptidyltransferase activity correlates with increased programmed −1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA 9:982–992
Noller HF, Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Lancaster L, Dallas A, Fredrick K, Earnest TN, Cate JH (2001) Structure of the ribosome at 5.5Å resolution and its interactions with functional ligands. Cold Spring Harb Symp Quant Biol 66:57–66
Oakes M, Aris JP, Brockenbrough JS, Wai H, Vu L, Nomura M (1998) Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J Cell Biol 143:23–34
Ogle JM, Ramakrishnan V (2005) Structural insights into translational fidelity. Annu Rev Biochem 74:129–177
Ohtake Y, Wickner RB (1995a) KRB1, a suppressor of mak7-1 (a mutant RPL4A), is RPL4B, a second ribosomal protein L4 gene, on a fragment of Saccharomyces chromosome XII. Genetics 140:129–137
Ohtake Y, Wickner RB (1995b) Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol Cell Biol 15:2772–2781
Pestka S (1977) Inhibitors of protein synthesis. In: Weissbach H, Pestka S (eds) Molecular mechanisms of protein biosynthesis. Academic, New York, pp 467–553
Petes TD (1979a) Meiotic mapping of yeast ribosomal deoxyribonucleic acid on chromosome XII. J Bacteriol 138:185–192
Petes TD (1979b) Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci USA 76:410–414
Petrov A, Meskauskas A, Dinman JD (2004) Ribosomal protein L3: influence on ribosome structure and function. RNA Biol 1:59–65
Rose MD, Winston F, Hieter P (1990) Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning, a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sigmund CD, Ettayebi M, Borden A, Morgan EA (1988) Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol 164:673–690
Smith MW, Meskauskas A, Wang P, Sergiev PV, Dinman JD (2001) Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol Cell Biol 21:8264–8275
Sommer SS, Wickner RB (1982) Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol 150:545–551
Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107:373–386
Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23:1008–1019
Steitz TA, Moore PB (2003) RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci 28:411–418
Stern S, Moazed D, Noller HF (1988) Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol 164:481–489
Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J (2002) 5S Ribosomal RNA Database. Nucleic Acids Res 30:176–178
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Velichutina IV, Hong JY, Mesecar AD, Chernoff YO, Liebman SW (2001) Genetic interaction between yeast Saccharomyces cerevisiae release factors and the decoding region of 18S rRNA. J Mol Biol 305:715–727
Wai HH, Vu L, Oakes M, Nomura M (2000) Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res 28:3524–3534
Wickner RB (1986) Double-stranded RNA replication in the yeast: the killer system. Annu Rev Biochem 55:373–395
Wickner RB (1996) Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev 60:250–265
Wickner RB, Leibowitz MJ (1976) Two chromosomal genes required for killing expression in killer strains of Saccharomyces cerevisiae. Genetics 82:429–442
Wickner RB, Porter-Ridley S, Fried HM, Ball SG (1982) Ribosomal protein L3 is involved in replication or maintenance of the killer double-stranded RNA genome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 79:4706–4708
Wilson DN, Nierhaus KH (2003) The ribosome through the looking Glass. Angew Chem Int Ed Engl 42:3464–3486
Xiong Y, Sundaralingam M (2000) Two crystal forms of helix II of Xenopus laevis 5S rRNA with a cytosine bulge. RNA 6:1316–1324
Yeh LC, Lee JC (1995) An in vitro system for studying RNA-protein interaction: application to a study of yeast ribosomal protein L1 binding to 5S rRNA. Biochimie 77:167–173
Yonath A et al (1998) Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry. Acta Crystallogr A 54:945–955
Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5Å resolution. Science 292:883–896
Acknowledgements
We wish to thank the members of the Dinman and Dontsova laboratories, with special thanks to Sarah Fraser, Ewan Plant, Steve Hutcheson and Alexey Bogdanov for their advice and support. This work was supported by grants to JDD from the National Institutes of Health (GM62143), and to JDD and OAD from the Fogarty International Center (TW005787), and to OAD from HHMI 55000303 and RFBR.
Author information
Authors and Affiliations
Corresponding author
Additional information
Sergey Kiparisov and Alexey Petrov contributed equally to this work
Rights and permissions
About this article
Cite this article
Kiparisov, S., Petrov, A., Meskauskas, A. et al. Structural and functional analysis of 5S rRNA in Saccharomyces cerevisiae . Mol Genet Genomics 274, 235–247 (2005). https://doi.org/10.1007/s00438-005-0020-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-0020-9