Abstract
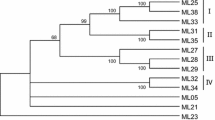
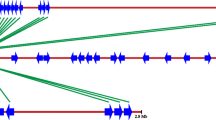
The scarcity of genetic polymorphism in Arachis hypogaea (peanut), as in other monophyletic polyploid species, makes it especially vulnerable to nematode, bacterial, fungal, and viral pathogens. Although no disease resistance genes have been cloned from peanut itself, the conserved motifs in cloned resistance genes from other plant species provide a means to isolate and analyze similar genes from peanut. To survey the number, diversity, evolutionary history, and genomic organization of resistance gene-like sequences in peanut, we isolated 234 resistance gene analogs (RGAs) by using primers designed from conserved regions of different classes of resistance genes including NBS-LRR, and LRR-TM classes. Phylogenetic and sequence analyses were performed to explore evolutionary relationships both among peanut RGAs and with orthologous genes from other plant taxa. Fifty-six overgos designed from the RGA sequences on the basis of their phyletic association were applied to a peanut BAC library; 736 hybridizing BAC clones were fingerprinted and contigs were formed in order to gain insights into the genomic organization of these genes. All the fingerprinting gels were blotted and screened with the respective overgos in order to verify the authenticity of the hits from initial screens, and to explore the physical organization of these genes in terms of both copy number and distribution in the genome. As a result, we identified 250 putative resistance gene loci. A correlation was found between the phyletic positions of the sequences and their physical locations. The BACs isolated here will serve as a valuable resource for future applications, such as map-based cloning, and will help improve our understanding of the evolution and organization of these genes in the peanut genome.






Similar content being viewed by others
References
Aarts MG, Hekkert A, Baste L, Holub EB, Beynon JL, Stiekema WJ, Pereira A (1998) Identification of R-gene homologous DNA fragments genetically linked to disease resistance loci in Arabidopsis thaliana. Mol Plant Microbe Interact 11:251–258
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anderson PA, Lawrence GJ, Morrish BC, Ayliffe MA, Finnegan EJ, Ellis JG (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9:641–651
Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ (1994) RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science 265:1856–1860
Bertioli DJ, Leal-Bertioli SCM, Lion MB, Santos VL, Pappas G, Cannon SB, Guimaraes PM (2003) A large scale analysis of resistance gene homologues in Arachis. Mol Genet Genomics 270:34–45
Blanc G, Wolfe KH (2004a) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Blanc G, Wolfe KH (2004b) Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16:1667–1678
Branch WD, Brenneman TB (1999) Stem rot disease evaluation of mass-selected peanut populations. Crop Prot 18:127–130
Burow MD, Simpson CE, Starr JL, Paterson AH (2001) Transmission genetics of chromatin from a synthetic amphidiploid to cultivated peanut (Arachis hypogaea L.): broadening the gene pool of a monophyletic polyploid species. Genetics 159:823–837
Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and Non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54:548–562
Cannon SB, McCombie WR, Sato S, Tabata S, Denny R, Palmer L, Katari M, Young ND, Stacey G (2003) Evolution and microsynteny of the apyrase gene family in three legume genomes. Mol Genet Genomics 270:347–361
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Di Gaspero G, Cipriani K (2003) Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to diseases resistance in grapevine. Mol Genet Genomics 269:612–623
Dixon MS, Hatzixanthis K, Jones DA, Harrison K, Jones JDG (1998) The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10:1915–1926
Ellis JG, Lawrence GJ, Luck JE, Dodds PN (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11:495–506
Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dang JL (1995) Structure of the Arabidopsis RPM1 gene enabling dual specific disease resistance. Science 269:843–846
Huettel B, Santra D, Muehlbauer J, Kahl G (2002) Resistance gene analogues of chickpea (Cicer arietinum L.): isolation, genetic mapping and association with a Fusarium resistance gene cluster. Theor Appl Genet 105:479–490
Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG (1994) Isolation of tomato Cf9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266:789–793
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kawchuk LM, Hachey J, Lynch DR, Kulcsar F, van Rooijen G, Waterer DR, Robertson A, Kokko E, Byers R, Howard RJ, Fischer R, Prufer D (2001) Tomato Ve disease resistance genes encode cell surface-like receptors. Proc Natl Acad Sci USA 98:6511–6515
Ku H-M, Vision T, Liu J, Tanksley SD (2000) Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci USA 97:9121–9126
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
Lagudah ES, Seah S, Sivasithamparam K, Karakousis A (1998) Cloning and characterization of a family of disease resistance gene analogs from wheat and barley. Theor Appl Genet 97:937–945
Lawrence GJ, Finnegan EJ, Ayliffe MA, Ellis JG (1995) The L6 gene for flax rust resistance is related to the Arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 7:1195–1206
Leung H, Line RF, Chen M (1998) Genome scanning for resistance-gene analogs in rice, barley, and wheat by high resolution electrophoresis. Theor Appl Genet 97:345–355
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1436
Meyers BC, Michelmore RW (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Meyers BC, Shen KA, Rohani P, Gaut BS, Michelmore RW (1998) Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10:1833–1846
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–322
Meyers BKA, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore R (2000) Genomic approaches to plant disease resistance. Curr Opin Plant Biol 3:125–131
Milligan SB, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson VM (1998) The Root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10:1307–1319
Nelson SC, Simpson CE, Starr JL (1989) Resistance to Meloidogyne arenaria in Arachis spp. germplasm. J Nematol (Suppl) 21:654–660
Ohmori T, Murata M, Motoyoshi F (1998) Characterization of disease resistance gene-like sequences in near-isogenic lines of tomato. Theor Appl Genet 96:331–338
Pan Q, Wendel J, Fluhr R (2000) Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J Mol Evol 50:203–213
Parniske M, Jones JDG (1999) Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc Natl Acad Sci USA 96:5850–5855
Pensuk V, Jogloy S, Wongkaew S, Patanothai A (2004) Generation means analysis of resistance to peanut bud necrosis caused by peanut bud necrosis tospovirus in peanut. Plant Breed 123:90–92
Penuela SD, Danesh D, Young ND (2002) Targeted isolation, sequence analysis, and physical mapping on nonTIR-NBS-LRR genes in soybean. Theor Appl Genet 104:261–272
Phipps PM, Porter DM (1998) Collar rot of peanut caused by Lasiodiplodia theobromae. Plant Dis 82:1205–1209
Richly E, Kurth J, Leister D (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19:76–84
Ross MT, LaBrie T, McPherson J, Stanton VM (1999) Screening large-insert libraries by hybridization. Wiley, New York, NY
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Saraste M, Sibbald PR, Wittinghofer A (1990) The P-Loop—a common motif in ATP-binding and GTP-binding proteins. Trends Biochem Sci 15:430–444
Seah S, Sivasithamparam K, Karakousis A, Lagudah ES (1998) Cloning and characterization of a family of disease resistance gene analogs from wheat and barley. Theor Appl Genet 97:937–945
Simillion C, Vandepoele K, Van Montagu MCE, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99:13627–13632
Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, Van der Lee T, Bleeker M, Onstenk J, de Both M, Haring M, Mes J, Cornelissen B, Zabeau M, Vos P (1998) Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 10:1055–1068
Soderlund C, Humphray S, Dunham A, French L (2000) Contigs built with fingerprints, markers, and FPC V4.7. Genome Res 10:1772–1787
Song WY, Wang GL, Shen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald PC (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:1804–1806
Sulston J, Mallett F, Durvin R, Horsnell T (1989) Image analysis of restriction enzyme fingerprint audioradiograms. Comput Appl Biosci 5:101–106
Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Traut TW (1994) The functions and consensus motifs of nine types of peptide segments that form different types of nucleotide-binding sites. Eur J Biochem 222:9–19
Vallad G, Rivkin M, Vallejos CE, McClean P (2001) Cloning and homology modelling of a Pto-like protein kinase family of common bean (Phaseolus vulgaris L.). Theor Appl Genet 103:1046–1058
Whitham S, McCormick S, Baker B (1996) The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc Natl Acad Sci USA 93:8776–8781
Yu Y, Buss G, Maroof M (1996) Isolation of a superfamily of candidate resistance genes in soybean based on a conserved nucleotide binding site. Proc Natl Acad Sci USA 93:11751–11756
Zhu HY, Cannon SB, Young ND, Cook DR (2002) Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol Plant Microbe Interact 15:529–539
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Hirsch
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yuksel, B., Estill, J.C., Schulze, S.R. et al. Organization and evolution of resistance gene analogs in peanut. Mol Genet Genomics 274, 248–263 (2005). https://doi.org/10.1007/s00438-005-0022-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-0022-7