Abstract
Conserved synteny––the sharing of at least one orthologous gene by a pair of chromosomes from two species––can, in the strictest sense, be viewed as sequence conservation between chromosomes of two related species, irrespective of whether coding or non-coding sequence is examined. The recent sequencing of multiple vertebrate genomes indicates that certain chromosomal segments of considerable size are conserved in gene order as well as underlying non-coding sequence across all vertebrates. Some of these segments lost genes or non-coding sequence and/or underwent breakage only in teleost genomes, presumably because evolutionary pressure acting on these regions to remain intact were relaxed after an additional round of whole genome duplication. Random reporter insertions into zebrafish chromosomes combined with computational genome-wide analysis indicate that large chromosomal areas of multiple genes contain long-range regulatory elements, which act on their target genes from several gene distances away. In addition, computational breakpoint analyses suggest that recurrent evolutionary breaks are found in “fragile regions” or “hotspots”, outside of the conserved blocks of synteny. These findings cannot be accommodated by the random breakage model and suggest that this view of genome and chromosomal evolution requires substantial reassessment.
Similar content being viewed by others
Introduction: the controversy
Twenty-three years ago Nadeau and Taylor (Nadeau and Taylor 1984) published an influential theoretical paper in which the conclusion was drawn that evolutionary chromosomal breakpoints that occurred since the divergence of human and mouse are distributed at random throughout the two genomes, thus leading to the now common view that conserved gene order on chromosomes of different species is a mere vestige of common ancestry without any functional implications. Their argument was one of statistical parsimony, stating that, since the lengths of discovered conserved chromosomal segments fitted a random distribution of evolutionary breaks, the postulate that certain chromosomal areas could not be broken was not necessary. Nadeau and Taylor’s paper concludes with “As a result, evidence other than linkage conservation in a few species is required to show that particular autosomal segments have been protected from rearrangement”. With the advent of multiple sequenced genomes, vertebrate chromosome evolution has become an area of increasing resolution and of many novel findings that require modification of conceptual frameworks evolved over the past 80 years. While few will contest that present day vertebrate genomes have arisen from an ancestral form through 2–3 successive rounds of whole genome duplications, as postulated by Ohno (1973) (e.g. Jaillon et al. 2004; Britten 2006), it is the rediploidization, which happened after these events, that is currently the topic of heated debates. Recently, the complete sequence of the human and mouse genomes have permitted an unprecedented view at vertebrate genomic architecture (Lander et al. 2001; Waterston et al. 2002) and with it the resolution of most gene neighborhoods, including novel gene predictions, and massive numbers of RNA genes and conserved non-coding elements (e.g. ENCODE_Project_Consortium 2007; Mikkelsen et al. 2007). These data were not available to Nadeau and Taylor (1984) when they calculated the lengths of chromosomal segments conserved since divergence of mouse and man. Instead, they relied on conserved gene order as markers on mouse/human linkage maps. They estimated the average length of conserved segments to be 8 cM. This number was then used to estimate the number of chromosomal rearrangements that have disrupted conserved linkage in the two genomes. However, this analysis could not take into account interruptions within these segments that do not result in changes in synteny (gene order). Therefore, by necessity, this analysis overestimated the length of conserved segments and, consequently, underestimated the number of rearrangements that occurred since the divergence of mouse and human. Post-genomic analyses indicate that, at the sequence level, 344 blocks of conserved synteny >100 kb can be discerned in human–mouse comparisons, and that these blocks are fragmented by many smaller rearrangements (Kent et al. 2003), suggesting that some assumptions upon which the Nadeau-Taylor calculations were based were erroneous.
Definitions
Before delving further into how the data compare then and now, it is important to make a few distinctions about the different levels of resolution in linkage maps and genome sequences.
-
Gene order: The traditional way of looking at conservation of chromosomal segments is through the order in which genes appear on chromosomes of two different species, and this was originally done using linkage maps. It is important to note that genes, as they appear on linkage maps, are usually one-dimensional markers, that is, they have no length, while in the genome sequence they do. The largest human gene, dystrophin, is larger than two million bases, and genes on the order of half a million bases are not unusual. Thus, at the sequence level, a gene is a conserved chromosomal segment.
-
Synteny: It denotes the presence of two or more genes on the same chromosome, and conserved synteny is the presence of two or more genes on one chromosome of each of two different species. Because the investigation of conserved synteny between mouse and human could originally only scrutinize conserved gene order, this has become the de facto meaning of conserved synteny. However, at the base pair resolution afforded by complete genome sequences, it becomes clear that genes are conserved in exonic as well as intronic sequence and often have extended regions of conserved non-coding sequence around them. Thus, in the classical sense (resulting from the use of a linkage map to measure conservation of synteny), a single gene, even if it spans 2 Mb, would not count as a region of synteny, even though a sequence of this length is largely conserved across species in exons as well as introns. Neither would a gene desert, an (sometimes very) extensive non-coding (conserved and non-conserved) region of a chromosome be considered syntenic between two species, even though it is clear that gene deserts resist chromosomal rearrangements and large chromosomal segments, even if containing only a single gene, are clearly conserved (Ovcharenko et al. 2005), nor would any other segment of any length containing no genes be considered to have conserved synteny, since it would appear as a segment with no markers on a conventional linkage map. Since both coding and non-coding sequence can be used as markers, we will use the term “conserved synteny” for any conserved sequence block, regardless of whether it encompasses multiple genes, an area containing a single gene, or areas devoid of known genes, as long as there is conservation at the sequence level.
Insights from genome-wide data: breakpoint reuse and breakpoint-resistant regions
In addition to the 344 long blocks of conserved synteny found by Kent et al. (2003), which roughly agree with the random breakage model, there are numerous short conserved chromosomal segments that do not agree with this assumption. Furthermore, Pevzner and Tesler (2003) found accumulation of evolutionary breakpoints in certain chromosomal segments termed “fragile” regions that are the sites of frequent “breakpoint reuse” in evolution, another fact that cannot be explained by the random breakage model (the “reused” sites in this case are not defined at the nucleotide level, but rather represent large genomic regions of up to several megabases where multiple evolutionary breakpoints have occurred independently (Pevzner and Tesler 2003) and in different mammalian lineages). This led to the formulation of the fragile breakage model (Pevzner and Tesler 2003). The reason why there should be solid and fragile chromosomal regions, however, was not understood and cannot be answered by genome inspection alone. The controversy continued with a paper (Sankoff and Trinh 2005) that attempted to rebuke the fragile breakage model by demonstrating that Pevzner and Tessler’s reasoning was based on artifacts caused by microrearrangements and imperfect algorithms for the determination of synteny blocks. The authors of the fragile breakage model recently struck back (Peng et al. 2006) by showing that Sankoff and Trinh’s alleged demonstration of the artifactual nature of fragile regions is a result of their deeply flawed synteny identification algorithm––which, if fixed, would leave them without their argument. At the same time, independent studies (Hinsch and Hannenhalli 2006; Ruiz-Herrera et al. 2006) provided additional evidence for breakpoint reuse and regions of apparent fragility across mammalian genomes.
Why would there be “evolutionary hotspots” in vertebrate genomes, and why are there apparent “solid” regions? The answer comes in two parts:
-
First, as proposed by Ohno (Ohno 1973), the proportion of loci involved in adaptive radiation is probably small, due to the evolutionary cost of eliminating unfit mutations at many loci simultaneously.
-
Second, there are loci that cannot be changed since their function is vital to embryonic development of all vertebrates.
The past several years have provided us with detailed view of the extent of the latter loci and their function. It has been noted that so-called developmental regulatory genes (encoding transcription factors, microRNAs, growth factors, receptors and other developmental regulators) are often found in gene-poor regions of the genome, and that many of them are also in the company of highly conserved non-coding elements spanning large areas around these genes and that these regions are conserved across species (Bejerano et al. 2004; Sandelin et al. 2004; Woolfe et al. 2005). Many of these elements have since been shown to be functional enhancers in reporter assays (de la Calle-Mustienes et al. 2005; Pennacchio et al. 2006). The fact that these elements sometimes act over distances exceeding hundreds of kilobases must mean that these elements and their target genes have to stay together in evolution, exerting strong selective pressure against breaks and any other rearrangements that would sever those elements from their target genes.
Kikuta et al. (2007) have recently demonstrated that certain genomic areas, which they termed genomic regulatory blocks (GRBs), are in fact regulatory domains that often serve the regulation of a single developmental gene (the target gene). Other genes in the area (the bystander genes) harbor the regulatory information (most often in their introns) necessary for correct expression of the target gene, resulting in effect in interlocked chains of genes that cannot be broken without serious loss of fitness (due to a loss of a large number of regulatory inputs to the target gene at once). This was shown by generating random insertions of reporter constructs into the zebrafish genome with the result that some of them would take on the expression pattern of a developmental gene far away, even though the insertion had occurred into an unrelated (bystander) gene. In each case where this was observed, the region surrounding the target gene was found to have conserved synteny throughout all sequenced vertebrate genomes. More of the same findings will undoubtedly turn up shortly when more highly conserved noncoding elements (HCNEs) are tested in reporter assays and the expression pattern driven by the enhancer is not that of the gene in (or near) which this sequence was found. Many of the target genes are so fundamental to vertebrate development that loss of a part of their regulatory input results in serious developmental anomalies or genetic disease.
The target gene in a GRB is often straightforward to predict. In the simplest case, a GRB consists of a gene desert dotted with HCNEs harboring only a single gene (Nobrega et al. 2003). In other cases there are, however, multiple genes in the region, including the aforementioned bystander genes. Even then, the target gene is usually detectable as coinciding with the highest density of noncoding conservation in the region, and by belonging to a restricted number of gene families. Less common, more complicated cases involve GRBs with multiple target genes, such as Iroquois clusters or DLX bigenes. In some other cases, the target genes, for instance microRNAs, may yet have to be discovered.
Integration
Although it is clear that more HCNEs will have to be tested for their regulatory activity and correlated to expression patterns of nearby genes, it would appear that the “solid” regions of Pevzner and Tesler (2003) correspond to a large extent to the GRBs of Kikuta et al. (2007). This would leave the fragile regions as those where evolutionary chromosomal breakpoints are possible, and where they could accumulate without harming the genetic integrity of the organism. It is, however, not yet known whether fragile regions are nevertheless what their name implies, namely intrinsically prone to breakage. For instance, it is not clear why certain genomic regions are recurrently broken in many cancers (Murphy et al. 2005), even though a number of associations with known genomic elements were proposed, including repeats (Ruiz-Herrera et al. 2006) and propensity for forming distinct secondary structures (Chuzhanova et al. 2003). What is evident is that GRBs need to keep their integrity and that evolution involving the target genes in them would have to proceed through small chromosomal alterations rather than wholesale rearrangements. The genomes of teleost fish reveal an interesting possible evolutionary pathway out of this “unbreakability” of GRBs: in them, a whole genome duplication (about 250 Myr ago) resulted in two copies of GRBs per haploid genome, each of which subsequently had a “window of opportunity” with increased freedom to mutate and break as long as each regulatory input remains functional in at least one of the copies. Various GRBs used this window of opportunity in different ways (Mulley et al. 2006; Kikuta et al. 2007).
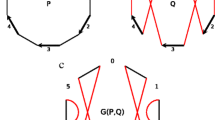
The different tolerance of neighborhoods of individual genes to breakpoints is therefore directly related to their long-range regulatory content. This in turn means that different genes differ greatly in the span of sequence from which they receive their regulatory input. Unlike the target genes of HCNEs, many other types of genes contain all their regulatory elements relatively close to the gene itself, making them tolerant to nearby breakages and rearrangements. Indeed, there are entire dense clusters of gene with little or no conserved synteny outside mammals, even though their 1-to-1 orthologs are present in evolutionarily more distant genomes. A schematic view of this notion is shown in Fig. 1.
Random breakage vs. genomic regulatory blocks. a The random breakage model assumes that, provided the breaks are not within genes, they can occur anywhere in the genome. In practice this means that any intergenic region is equally susceptible to evolutionary breaks. b Genomic regulatory blocks, supported by cross-species comparisons and reporter insertions (Kikuta et al. 2007) imply that a particular gene (target gene) can receive regulatory inputs from large genomic regions that contain other, unrelated genes (bystander genes). A break between target and bystander gene in a GRB would result in partial loss of regulatory input to the target gene, and is therefore actively selected against
The question that remains open is that of apparent breakpoint reuse (Pevzner and Tesler 2003; Hinsch and Hannenhalli 2006), which would imply the existence of regions of chromosomally increased fragility. It is unknown whether these regions are physically more fragile, or merely represent locations where selection against breakpoints is minimal. Peng et al. (2006) attempted to associate the breakpoint reuse with the size of “upstream regulatory regions” that they attached to genes in their model, and saw an increase in apparent breakpoint reuse with the increasing size of that region. They concluded that “long regulatory regions and inhomogeneity of gene distribution in mammalian genomes may provide at least partial explanation for the fragile breakpoint model”. It is intuitively plausible that using GRBs in place of upstream regions in the above model would increase the overall proportion of “unbreakable regions” in the genome and would therefore account for an even higher proportion of the observed breakpoint reuse. Due to current difficulties in defining GRBs in a formal way, a definitive computational demonstration of this phenomenon remains an exciting open problem.
Conclusion
Long-range regulation and the transformation of the concept of a gene (Gerstein et al. 2007) have provided us with a definite nail in the coffin for Nadeau and Taylor’s hypothesis. While it is still unclear why some evolutionary breakpoints are apparently reused, the evidence for multigene regions in which breakpoints are not tolerated is both strong and clear. Regions of long-range regulation with multiple genes intertwined with regulatory elements controlling neighboring genes have emerged as functional units in the genome that are “protected from rearrangements” simply through the fact that breaks in them lead to target gene dysregulation that results in reduced fitness, and will therefore not be passed on in the gene pool.
References
Bejerano G et al (2004) Ultraconserved elements in the human genome. Science 304:1321–1325
Britten RJ (2006) Almost all human genes resulted from ancient duplication. Proc Natl Acad Sci USA 103:19027–19032
Chuzhanova N, Abeysinghe SS, Krawczak M, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer II: potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum Mutat 22:245–251
de la Calle-Mustienes E et al (2005) A functional survey of the enhancer activity of conserved non-coding sequences from vertebrate Iroquois cluster gene deserts. Genome Res 15:1061–1072
ENCODE_Project_Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816
Gerstein MB et al (2007) What is a gene, post-ENCODE? History and updated definition. Genome Res 17:669–681
Hinsch H, Hannenhalli S (2006) Recurring genomic breaks in independent lineages support genomic fragility. BMC Evol Biol 6:90
Jaillon O et al (2004) Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957
Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D (2003) Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA 100:11484–11489
Kikuta H et al (2007) Genomic regulatory blocks encompass multiple neighboring genes and maintain conserved synteny in vertebrates. Genome Res 17:545–555
Lander ES et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Mikkelsen TS et al (2007) Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447:167–177
Mulley JF, Chiu CH, Holland PW (2006) Breakup of a homeobox cluster after genome duplication in teleosts. Proc Natl Acad Sci USA 103:10369–10372
Murphy WJ et al (2005) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309:613–617
Nadeau JH, Taylor BA (1984) Lengths of chromosomal segments conserved since divergence of man and mouse. Proc Natl Acad Sci USA 81:814–818
Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413
Ohno S (1973) Ancient linkage groups and frozen accidents. Nature 244:259–262
Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L (2005) Evolution and functional classification of vertebrate gene deserts. Genome Res 15:137–145
Peng Q, Pevzner PA, Tesler G (2006) The fragile breakage versus random breakage models of chromosome evolution. PLoS Comput Biol 2:e14
Pennacchio LA et al (2006) In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499–502
Pevzner P, Tesler G (2003) Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA 100:7672–7677
Ruiz-Herrera A, Castresana J, Robinson TJ (2006) Is mammalian chromosomal evolution driven by regions of genome fragility? Genome Biol 7:R115
Sandelin A et al (2004) Arrays of ultraconserved non-coding regions span the loci of key developmental genes in vertebrate genomes. BMC Genomics 5:99
Sankoff D, Trinh P (2005) Chromosomal breakpoint reuse in genome sequence rearrangement. J Comput Biol 12:812–821
Waterston RH et al (2002) Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Woolfe A et al (2005) Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3:e7
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Van de Peer.
Rights and permissions
About this article
Cite this article
Becker, T.S., Lenhard, B. The random versus fragile breakage models of chromosome evolution: a matter of resolution. Mol Genet Genomics 278, 487–491 (2007). https://doi.org/10.1007/s00438-007-0287-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0287-0