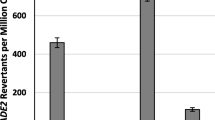
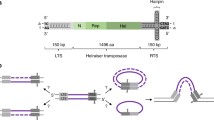
Abstract
Previous studies have shown that the transposase and the inverted terminal repeat (ITR) of the Mos1 mariner elements are suboptimal for transposition; and that hyperactive transposases and transposon with more efficient ITR configurations can be obtained by rational molecular engineering. In an attempt to determine the extent to which this element is suboptimal for transposition, we investigate here the impact of the three main DNA components on its transposition efficiency in bacteria and in vitro. We found that combinations of natural and synthetic ITRs obtained by systematic evolution of ligands by exponential enrichment did increase the transposition rate. We observed that when untranslated terminal regions were associated with their respective natural ITRs, they acted as transposition enhancers, probably via the early transposition steps. Finally, we demonstrated that the integrity of the Mos1 inner region was essential for transposition. These findings allowed us to propose prototypes of optimized Mos1 vectors, and to define the best sequence features of their associated marker cassettes. These vector prototypes were assayed in HeLa cells, in which Mos1 vectors had so far been found to be inactive. The results obtained revealed that using these prototypes does not circumvent this problem. However, such vectors can be expected to provide new tools for the use in genome engineering in systems such as Caenorhabditis elegans in which Mos1 is very active.







Similar content being viewed by others
Abbreviations
- HTH:
-
Helix-turn-helix
- ITR:
-
Inverted terminal repeat
- ORF:
-
Open reading frame
- SELEX:
-
Systematic evolution of ligands by exponential enrichment
- TE:
-
Transposable element
- UTR:
-
Untranslated terminal region
References
Augé-Gouillou C, Hamelin MH, Demattei MV, Periquet G, Bigot Y (2001a) The ITR binding domain of the Mariner Mos-1 transposase. Mol Genet Genomics 265:58–65
Augé-Gouillou C, Hamelin MH, Demattei MV, Periquet M, Bigot Y (2001b) The wild-type conformation of the Mos1 inverted terminal repeats is suboptimal for transposition in bacteria. Mol Genet Genomics 265:51–57
Augé-Gouillou C, Brillet B, Hamelin MH, Bigot Y (2005) Assembly of the mariner Mos1 synaptic complex. Mol Cell Biol 25:2861–2870
Bessereau JL, Wright A, Williams DC, Schuske K, Davis MW, Jorgensen EM (2001) Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413:70–74
Bigot Y, Brillet B, Augé-Gouillou C (2005) Conservation of palindromic and mirror motifs within inverted terminal repeats of mariner-like elements. J Mol Biol 351:108–116
Boulin T, Bessereau JL (2007) Mos1-mediated insertional mutagenesis in Caenorhabditis elegans. Nat Protoc 2:1276–1287
Brillet B, Bigot Y, Augé-Gouillou C (2007) Assembly of the Tc1 and mariner transposition initiation complexes depends on the origins of their transposase DNA binding domains. Genetica 130:105–120
Coates CJ, Jasinskiene N, Miyashiro L, James AA (1998) Mariner transposition and transformation of the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci USA 95:3748–3751
Crénès G, Ivo D, Hérisson J, Dion S, Renault S, Bigot Y, Petit A (2009) The bacterial Tn9 chloramphenicol resistance gene: an attractive DNA segment for Mos1 mariner insertions. Mol Genet Genomics 281:315–328
Dawson A, Finnegan DJ (2003) Excision of the Drosophila mariner transposon Mos1: comparison with bacterial transposition and V(D)J recombination. Mol Cell 11:225–235
Derbyshire KM, Hatfull G, Willetts N (1987) Mobilization of the non-conjugative plasmid RSF1010: a genetic and DNA sequence analysis of the mobilization region. Mol Genet Genomics 206:161–168
Fadool JM, Hartl DL, Dowling JE (1998) Transposition of the mariner element from Drosophila mauritiana in zebrafish. Proc Natl Acad Sci USA 95:5182–5186
Fischer SE, Wienholds E, Plasterk RH (2001) Regulated transposition of a fish transposon in the mouse germ line. Proc Natl Acad Sci USA 98:6759–6764
Gabrielian A, Pongor S (1996) Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett 393:65–68
Gabrielian A, Simoncsits A, Pongor S (1996) Distribution of bending propensity in DNA sequences. FEBS Lett 393:124–130
Gabrielian A, Vlahovicek K, Pongor S (1997) Distribution of sequence-dependent curvature in genomic DNA. FEBS Lett 406:69–74
Germon S, Casteret S, Carpentier G, Bouchet N, Adet J, Bigot Y, Augé-Gouillou C (2009) Transposase optimization of the Mos1 mariner transposon by rational mutagenesis. Genetica (in press)
Goodsell DS, Dickerson RE (1994) Bending and curvature calculations in B-DNA. Nucleic Acids Res 22:5497–5503
Goyard S, Tosi LR, Gouzova J, Majors J, Beverley SM (2001) New Mos1 mariner transposons suitable for the recovery of gene fusions in vivo and in vitro. Gene 280:97–105
Gueiros-Filho FJ, Beverley SM (1997) Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716–1719
Halaimia-Toumi N, Casse N, Demattei MV, Renault S, Pradier E, Bigot Y (2004) The GC-richt Bytmar1 from the deep-Sea hydrothermal crab, Bythograea thermydron, may encode three transposase isoforms from a single ORF. J Mol Evol 59:747–760
Hérisson J, Ferey N, Gros PE, Gherbi R (2007) ADN-viewer: a 3D approach for bioinformatic analyses of large DNA sequences. Cell Mol Biol 52:24–31
Keravala A, Liu D, Lechman ER, Wolfe D, Nash JA, Lampe DJ, Robbins PD (2006) Hyperactive Himar1 transposase mediates transposition in cell culture and enhances gene expression in vivo. Hum Gene Ther 17:1006–1018
Kidwell MG, Lisch DR (2001) Perspective: transposable elements, parasitic DNA and genome evolution. Evolution 55:1–24
Lampe DJ, Grant TE, Robertson HM (1998) Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179–187
Levine HA, Nilsen-Hamilton M (2007) A mathematical analysis of SELEX. Comput Biol Chem 31:11–35
Lohe AR, Hartl DL (2002) Efficient mobilization of mariner in vivo requires multiple internal sequences. Genetics 160:519–526
Lohe AR, Timmons C, Beerman I, Lozovskaya ER, Hartl DL (2000) Self-inflicted wounds, template-directed gap repair and a recombination hotspot: effects of the mariner transposase. Genetics 154:647–656
Lozovsky ER, Nurminsky D, Wimmer EA, Hartl DL (2002) Unexpected stability of mariner transgenes in Drosophila. Genetics 160:527–535
Naumann TA, Reznikoff WS (2000) Trans catalysis in Tn5 transposition. Proc Natl Acad Sci USA 97:8944–8949
Naumann TA, Reznikoff WS (2002) Tn5 Transposase with an altered specificity for transposon ends. J Bact 184:233–240
Neumann U, Khalaf H, Rimpler M (1994) Quantitation of electrophoretically separated proteins in the submicrogram range by dye elution. Electrophoresis 15:916–921
Pledger DW, Fu YQ, Coates CJ (2004) Analyses of cis -acting elements that affect the transposition of Mos1 mariner transposons in vivo. Mol Genet Genomics 272:67–75
Rengarajan K, Cristol SM, Mehta M, Nickerson JM (2002) Quantifying DNA concentrations using fluorometry: a comparison of fluorophores. Mol Vis 8:416–421
Richardson JM, Dawson A, O’Hagan N, Taylor P, Finnegan DJ, Walkinshaw MD (2006) Mechanism of Mos1 transposition: insights from structural analysis. EMBO J 25:1324–1334
Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H (1998) Transposition of the Drosophila element mariner into the chicken germ line. Nat Biotechnol 16:1050–1053
Sinzelle L, Jegot G, Brillet B, Rouleux-Bonnin F, Bigot Y, Augé-Gouillou C (2008) Factors acting on Mos1 transposition efficiency. BMC Mol Biol 9:106
Tosi LR, Beverley SM (2000) cis and trans factors affecting Mos1 mariner evolution and transposition in vitro, and its potential for functional genomics. Nucleic Acids Res 28:784–790
Wiegand TW, Reznikoff WS (1992) Characterization of two hypertransposing Tn5 mutants. J Bacteriol 174:1229–1239
Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM (2006) piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA 103:15008–15013
Zhang L, Dawson A, Finnegan DJ (2001) DNA-binding activity and subunit interaction of the mariner transposase. Nucl Acid Res 29:3566–3575
Acknowledgments
This work was supported by the University of François Rabelais of Tours, and funded by grants from the European Commission (Project SyntheGeneDelivery, N°018716), the C.N.R.S., the Ministère de l’Education Nationale, de la Recherche et de la Technologie, the Association Française contre la Myopathie, and the Groupement de Recherche CNRS 2157. The technical assistance of Sylvie Bigot is warmly acknowledged. The English text has been revised by Dr M. Ghosh.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Grandbastien.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Casteret, S., Chbab, N., Cambefort, J. et al. Physical properties of DNA components affecting the transposition efficiency of the mariner Mos1 element. Mol Genet Genomics 282, 531–546 (2009). https://doi.org/10.1007/s00438-009-0484-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-009-0484-0