Abstract
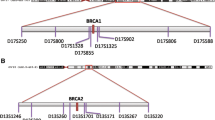
Three founder mutations in BRCA1 and BRCA2 contribute to the risk of hereditary breast and ovarian cancer in Ashkenazi Jews (AJ). They are observed at increased frequency in the AJ compared to other BRCA mutations in Caucasian non-Jews (CNJ). Several authors have proposed that elevated allele frequencies in the surrounding genomic regions reflect adaptive or balancing selection. Such proposals predict long-range linkage disequilibrium (LD) resulting from a selective sweep, although genetic drift in a founder population may also act to create long-distance LD. To date, few studies have used the tools of statistical genomics to examine the likelihood of long-range LD at a deleterious locus in a population that faced a genetic bottleneck. We studied the genotypes of hundreds of women from a large international consortium of BRCA1 and BRCA2 mutation carriers and found that AJ women exhibited long-range haplotypes compared to CNJ women. More than 50% of the AJ chromosomes with the BRCA1 185delAG mutation share an identical 2.1 Mb haplotype and nearly 16% of AJ chromosomes carrying the BRCA2 6174delT mutation share a 1.4 Mb haplotype. Simulations based on the best inference of Ashkenazi population demography indicate that long-range haplotypes are expected in the context of a genome-wide survey. Our results are consistent with the hypothesis that a local bottleneck effect from population size constriction events could by chance have resulted in the large haplotype blocks observed at high frequency in the BRCA1 and BRCA2 regions of Ashkenazi Jews.





Similar content being viewed by others
Abbreviations
- AJ:
-
Ashkenazi Jews
- CIMBA:
-
Consortium of Investigators of Modifiers of BRCA1 and BRCA2
- CNJ:
-
Caucasian non-Jews
- LD:
-
Linkage disequilibrium
- MAF:
-
Minor allele frequency
- MJ:
-
Median joining
- PCA:
-
Principal components analysis
- SNP:
-
Single nucleotide polymorphism
- IBD:
-
Identity by descent
References
Akey JM, Zhang G, Zhang K, Jin L, Shriver MD (2002) Interrogating a high-density SNP map for signatures of natural selection. Genome Res 12:1805–1814. doi:10.1101/gr.631202
Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, Ding YC, Rebbeck TR, Weitzel JN, Lynch HT, Isaacs C, Ganz PA, Tomlinson G, Olopade OI, Couch FJ, Wang X, Lindor NM, Pankratz VS, Radice P, Manoukian S, Peissel B, Zaffaroni D, Barile M, Viel A, Allavena A, Dall’olio V, Peterlongo P, Szabo CI, Zikan M, Claes K, Poppe B, Foretova L, Mai PL, Greene MH, Rennert G, Lejbkowicz F, Glendon G, Ozcelik H, Andrulis IL, Thomassen M, Gerdes AM, Sunde L, Cruger D, Birk Jensen U, Caligo M, Friedman E, Kaufman B, Laitman Y, Milgrom R, Dubrovsky M, Cohen S, Borg A, Jernstrom H, Lindblom A, Rantala J, Stenmark-Askmalm M, Melin B, Nathanson K, Domchek S, Jakubowska A, Lubinski J, Huzarski T, Osorio A, Lasa A, Duran M, Tejada MI, Godino J, Benitez J, Hamann U, Kriege M, Hoogerbrugge N, van der Luijt RB, Asperen CJ, Devilee P, Meijers-Heijboer EJ, Blok MJ, Aalfs CM, Hogervorst F, Rookus M, Cook M, Oliver C, Frost D, Conroy D, Evans DG, Lalloo F, Pichert G, Davidson R, Cole T, Cook J, Paterson J, Hodgson S, Morrison PJ, Porteous ME, Walker L, Kennedy MJ, Dorkins H, Peock S, Godwin AK, Stoppa-Lyonnet D, de Pauw A et al (2010) Common Breast Cancer Susceptibility Alleles and the Risk of Breast Cancer for BRCA1 and BRCA2 Mutation Carriers: Implications for Risk Prediction. Cancer Res 70:9742–9754. doi:10.1158/0008-5472.CAN-10-1907
Atzmon G, Hao L, Pe’er I, Velez C, Pearlman A, Palamara PF, Morrow B, Friedman E, Oddoux C, Burns E, Ostrer H (2010) Abraham’s children in the genome era: major Jewish diaspora populations comprise distinct genetic clusters with shared Middle Eastern Ancestry. Am J Hum Genet 86:850–859
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Bray SM, Mulle JG, Dodd AF, Pulver AE, Wooding S, Warren ST (2010) Signatures of founder effects, admixture, and selection in the Ashkenazi Jewish population. Proc Natl Acad Sci USA 107:16222–16227. doi:10.1073/pnas.1004381107
Carlson CS, Thomas DJ, Eberle MA, Swanson JE, Livingston RJ, Rieder MJ, Nickerson DA (2005) Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res 15:1553–1565
Chakravarti A, Chakraborty R (1978) Elevated frequency of Tay-Sachs disease among Ashkenazic Jews unlikely by genetic drift alone. Am J Hum Genet 30:256–261
Chenevix-Trench G, Milne RL, Antoniou AC, Couch FJ, Easton DF, Goldgar DE (2007) An international initiative to identify genetic modifiers of cancer risk in BRCA1 and BRCA2 mutation carriers: the Consortium of Investigators of Modifiers of BRCA1 and BRCA2 (CIMBA). Breast Cancer Res 9:104. doi:10.1186/bcr1670
Clayton D. SNPHAP: a program for estimating frequencies of large haplotypes of SNPs. http://www-gene.cimr.cam.ac.uk/clayton/software
Cochran G, Hardy J, Harpending H (2006) Natural history of Ashkenazi intelligence. J Biosoc Sci 38:659–693. doi:10.1017/S0021932005027069
Ewens WJ (1978) Tay-Sachs disease and theoretical population genetics. Am J Hum Genet 30:328–329
Gaudet MM, Kirchhoff T, Green T, Vijai J, Korn JM, Guiducci C, Segre AV, McGee K, McGuffog L, Kartsonaki C, Morrison J, Healey S, Sinilnikova OM, Stoppa-Lyonnet D, Mazoyer S, Gauthier-Villars M, Sobol H, Longy M, Frenay M, Collaborators GS, Hogervorst FB, Rookus MA, Collee JM, Hoogerbrugge N, van Roozendaal KE, Piedmonte M, Rubinstein W, Nerenstone S, Van Le L, Blank SV, Caldes T, de la Hoya M, Nevanlinna H, Aittomaki K, Lazaro C, Blanco I, Arason A, Johannsson OT, Barkardottir RB, Devilee P, Olopade OI, Neuhausen SL, Wang X, Fredericksen ZS, Peterlongo P, Manoukian S, Barile M, Viel A, Radice P, Phelan CM, Narod S, Rennert G, Lejbkowicz F, Flugelman A, Andrulis IL, Glendon G, Ozcelik H, Toland AE, Montagna M, D’Andrea E, Friedman E, Laitman Y, Borg A, Beattie M, Ramus SJ, Domchek SM, Nathanson KL, Rebbeck T, Spurdle AB, Chen X, Holland H, John EM, Hopper JL, Buys SS, Daly MB, Southey MC, Terry MB, Tung N, Overeem Hansen TV, Nielsen FC, Greene MI, Mai PL, Osorio A, Duran M, Andres R, Benitez J, Weitzel JN, Garber J, Hamann U, Peock S, Cook M, Oliver C, Frost D, Platte R, Evans DG, Lalloo F, Eeles R, Izatt L, Walker L, Eason J et al (2010) Common genetic variants and modification of penetrance of BRCA2-associated breast cancer. PLoS Genet 6:e1001183. doi:10.1371/journal.pgen.1001183
Gibson J, Morton NE, Collins A (2006) Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 15:789–795. doi:10.1093/hmg/ddi493
Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P (2005) Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res 15:790–799. doi:10.1101/gr.3541005
Hartl DL, Clark AG (2007) Principles of Population Genetics, Fourth edn edn. Sinauer Associates, Inc, Sunderland
Hellenthal G, Stephens M (2007) msHOT: modifying Hudson’s ms simulator to incorporate crossover and gene conversion hotspots. Bioinformatics 23:520–521
Hudson R (2002) Generating samples under a Wright-Fisher model of genetic variation. Bioinformatics 18:337–338
Huttley G, Easteal S, Southey M, Tesoriero A, Giles G, McCredie MRE, Hopper J, Venter D, Study ABCF (2000) Adapative evolution of the tumour suppressor BRCA1 in humans and chimpanzees. Nat Genet 25:410–413
King MC, Marks JH, Mandell JB (2003) Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science 3002:643–646
Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine-scale map of recombination rates and hotspots across the human genome. Science 310:247–248
Neuhausen S, Godwin A, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, Csokay B, Serova O, Lalloo F, Osorio A, Stratton M, Offit K, Boyd J et al (1998) Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet 62:1381–1388
Olshen AB, Gold B, Lohmueller KE, Struewing JP, Satagopan J, Stefanov SA, Eskin E, Kirchhoff T, Lautenberger JA, Klein RJ, Friedman E, Norton L, Ellis NA, Viale A, Lee CS, Borgen PI, Clark AG, Offit K, Boyd J (2008) Analysis of genetic variation in Ashkenazi Jews by high density SNP genotyping. BMC Genet 9:14
Ostrer H (2001) A genetic profile of contemporary Jewish populations. Nat Rev Genet 2:891–898
Pereira LHM, Pineda MA, Rowe WH, Fonseca LR, Greene M, Offit K, Ellis NA, Zhang J, Collins A, Struewing J (2007) The BRCA1 Ashkenazi founder mutations occur on common haplotypes and are not highly correlated with anonymous single nucleotide polymorphisms likely to be used in genome-wide case–control association studies. BMC Genet 8:68
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909. doi:10.1038/ng1847
Sabeti P, Reich D, Higgins J, Levine HZ, Richter D, Schaffner S, Gabriel S, Platko J, Patterson N, McDonald G, Ackerman H, Campbell S, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander E (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419:832–837
Saunders MA, Slatkin M, Garner C, Hammer MF, Nachman MW (2005) The extent of linkage disequilibrium caused by selection on G6PD in humans. Genetics 171:1219–1229
Slatkin M (2004) A population-genetic test of founder effects and implications for Ashkenazi Jewish disease. Am J Hum Genet 75:282–293
Slatkin M, Rannala B (2000) Estimating allele age. Annu Rev Genomics Hum Genet 1:225–249. doi:10.1146/annurev.genom.1.1.225
Stefanov S, Lautenberger J, Gold B (2008) An analysis pipeline for genome-wide association studies. Cancer Inform 6:455–461
Struewing JP, Hartge P, Wacholder S, Baker S, Berlin M, McAdams M, Timmerman M, Brody L, Tucker M (1997) The risk of cancer associated with specific mutations of BRCA1 and BRCA2. N Engl J Med 336:1401–1408
Struewing JP, Coriaty ZM, Ron E, Livoff A, Konichezky M, Cohen P, Resnick MB, Lifzchiz-Mercerl B, Lew S, Iscovich J (1999) Founder BRCA1/2 mutations among male patients with breast cancer in Israel. Am J Hum Genet 65:1800–1802. doi:10.1086/302678
Thorlacius S, Sigurdsson S, Bjarnadottir H, Olafsdottir G, Jonasson JG, Tryggvadottir L, Tulinius H, Eyfjord JE (1997) Study of a single BRCA2 mutation with high carrier frequency in a small population. Am J Hum Genet 60:1079–1084
Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt C, Silverman J, Powell K, Mortensen H, Hirbo J, Osman M, Ibrahim M, Omar S et al (2007) Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 39:31–40
Tonin P, Weber B, Offit K, Couch F, Rebbeck TR, Neuhausen S, Godwin A, Daly M, Wagner-Costalos J, Berman D, Grana G, Fox E et al (1996) Frequency of recurrent BRCA1 and BRCA2 mutations in Ashkenazi breast cancer families. Nat Med 2:1179–1183
Voight BF, Kudaravalli S, Wen X, Pritchard JK (2006) A Map of Recent Positive Selection in the Human Genome. PLoS Biology 4:e72
Wagener D, Cavalli-Sforza LL, Barakat R (1978) Ethnic variation of genetic disease: roles of drift for recessive lethal genes. Am J Hum Genet 30:262–270
Weir B, Cockerham C (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgments
This work was supported in part by federal funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. We also acknowledge the support of the Starr Foundation, the Breast Cancer Research Foundation and the Sabin Family Fund. The content of this publication does not necessarily reflect the views of the Department of Health and Human Services nor does its mention of trade names, commercial products or organizations imply endorsement by the US government. The authors wish to thank Dr. Colm O’hUigin, who provided valuable comments on an early version of this manuscript.
UKFOCR was supported by a project grant from CRUK to Paul Pharoah. We thank Paul Pharoah, Simon Gayther, Susan Ramus, Carole Pye and Patricia Harrington for their contributions toward the UKFOCR.
The GEMO study (Cancer Genetics Network “Groupe Génétique et Cancer”, Fédération Nationale des Centres de Lutte Contre le Cancer, France) is supported by the Ligue National Contre le Cancer; Association for International Cancer Research Grant (AICR-07-0454); and the Association “Le cancer du sein, parlons-en!” Award.
We wish to thank all the GEMO collaborating groups for their contribution to this study. GEMO Collaborating Centers are: Coordinating Centres, Unité Mixte de Génétique Constitutionnelle des Cancers Fréquents, Centre Hospitalier Universitaire de Lyon/Centre Léon Bérard, & UMR5201 CNRS, Université de Lyon, Lyon: Olga Sinilnikova, Laure Barjhoux, Sophie Giraud, Mélanie Léone, Sylvie Mazoyer; and INSERM U509, Service de Génétique Oncologique, Institut Curie, Paris: Dominique Stoppa-Lyonnet, Marion Gauthier-Villars, Claude Houdayer, Virginie Moncoutier, Muriel Belotti, Antoine de Pauw. Institut Gustave Roussy, Villejuif: Brigitte Bressac-de-Paillerets, Audrey Remenieras, Véronique Byrde, Olivier Caron, Gilbert Lenoir. Centre Jean Perrin, Clermont–Ferrand: Yves-Jean Bignon, Nancy Uhrhammer. Centre Léon Bérard, Lyon: Christine Lasset, Valérie Bonadona. Centre François Baclesse, Caen: Agnès Hardouin, Pascaline Berthet. Institut Paoli Calmettes, Marseille: Hagay Sobol, Violaine Bourdon, Tetsuro Noguchi, François Eisinger. Groupe Hospitalier Pitié-Salpétrière, Paris: Florence Coulet, Chrystelle Colas, Florent Soubrier. CHU de Arnaud-de-Villeneuve, Montpellier: Isabelle Coupier. Centre Oscar Lambret, Lille: Jean-Philippe Peyrat, Joëlle Fournier, Françoise Révillion, Philippe Vennin, Claude Adenis. Centre René Huguenin, St Cloud: Etienne Rouleau, Rosette Lidereau, Liliane Demange, Catherine Nogues. Centre Paul Strauss, Strasbourg: Danièle Muller, Jean-Pierre Fricker. Institut Bergonié, Bordeaux: Michel Longy, Nicolas Sevenet. Institut Claudius Regaud, Toulouse: Christine Toulas, Rosine Guimbaud, Laurence Gladieff, Viviane Feillel. CHU de Grenoble: Dominique Leroux, Hélène Dreyfus, Christine Rebischung. CHU de Dijon: Cécile Cassini, Laurence Faivre. CHU de St-Etienne: Fabienne Prieur. Hôtel Dieu Centre Hospitalier, Chambéry: Sandra Fert Ferrer. Centre Antoine Lacassagne, Nice: Marc Frénay. CHU de Limoges: Laurence Vénat-Bouvet. Creighton University, Omaha, USA: Henry T. Lynch.
For kConFab, we wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinic and the Clinical Follow Up Study (funded 2001–2009 by NHMRC and currently by the National Breast Cancer Foundation and Cancer Australia #628333) for their contributions to this resource, and the many families who contributed to kConFab. kConFab is supported by grants from the National Breast Cancer Foundation, the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia.
The CONSIT TEAM study (Consorzio degli Studi Italiani Tumori Ereditari Alla Mammella), acknowledges Marco Pierotti, Siranoush Manoukian, Daniela Zaffaroni, Carla B. Ripamonti and Paolo Radice of the Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy; Monica Barile of the Istituto Europeo di Oncologia, Milan, Italy and Loris Bernard of the Cogentech, Consortium for Genomic Technologies, Milan, Italy. We thank all patients and families who participated in this study. This study was supported by funds from Italian citizens who allocated the 5 × 1,000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1,000’).
For the Mayo Study, the authors would like to acknowledge funding from the Breast Cancer Research Foundation, Komen Foundation, and NIHCA116167.
The Cancer Prevention Institute of California was supported by the National Cancer Institute, National Institutes of Health under RFA-CA-06-503 and through cooperative agreements with members of the Breast Cancer Family Registry and P.I.s. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Cancer Family Registry (CFR), nor does mention of trade names, commercial products or organizations imply endorsement by the US government or the CFR.
The SWE-BRCA study acknowledge the study collaborators: Per Karlsson, Margareta Nordling, Annika Bergman and Zakaria Einbeigi, Gothenburg, Sahlgrenska University Hospital; Marie Stenmark-Askmalm and Sigrun Liedgren Linköping University Hospital; Åke Borg, Niklas Loman, Håkan Olsson, Ulf Kristoffersson, Helena Jernström, Katja Harbst and Karin Henriksson, Lund University Hospital; Annika Lindblom, Brita Arver, Anna von Wachenfeldt, Annelie Liljegren, Gisela Barbany-Bustinza and Johanna Rantala, Stockholm, Karolinska University Hospital; Beatrice Melin, Henrik Grönberg, Eva-Lena Stattin and Monica Emanuelsson, Umeå University Hospital; Hans Ehrencrona, Richard Rosenquist Brandell and Niklas Dahl, Uppsala University Hospital. The HCSC study was supported by RD06/0020/0021 from ISCIII.
The Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) Collaborating Centers are Coordinating center: Netherlands Cancer Institute, Amsterdam, NL: F.B.L. Hogervorst, S. Verhoef, M. Verheus, L.J. van ‘t Veer, F.E. van Leeuwen, M.A. Rookus; Erasmus Medical Center, Rotterdam, NL: M. Collée, A.M.W. van den Ouweland, A. Jager, M.J. Hooning, M.M.A. Tilanus-Linthorst, C. Seynaeve; Leiden University Medical Center, NL, Leiden: C.J. van Asperen, J.T. Wijnen, M.P. Vreeswijk, R.A. Tollenaar, P. Devilee; Radboud University Nijmegen Medical Center, Nijmegen, NL: M.J. Ligtenberg, N. Hoogerbrugge; University Medical Center Utrecht, Utrecht, NL: M.G. Ausems, R.B. van der Luijt; Amsterdam Medical Center, NL: C.M. Aalfs, T.A. van Os; VU University Medical Center, Amsterdam, NL: J.J.P. Gille, Q. Waisfisz, H.E.J. Meijers-Heijboer; University Hospital Maastricht, Maastricht, NL: E.B. Gomez-Garcia, C.E. van Roozendaal, Marinus J. Blok, B. Caanen; University Medical Center Groningen University, NL: J.C. Oosterwijk, A.H. van der Hout, M.J. Mourits; The Netherlands Foundation for the detection of hereditary tumours, Leiden, NL: H.F. Vasen. The HEBON study is supported by the Dutch Cancer Society grants NKI1998-1854, NKI2004-3088 and NKI2007-3756.
The Ohio State University Clinical Cancer Genetics (OSU CCG) study is supported by the OSU Comprehensive Cancer Center. We thank Leigha Senter and Kevin Sweet for patient accrual, sample ascertainment and database management. The Human Genetics Sample bank processed the samples.
This work was supported by Cancer Care Ontario and the US National Cancer Institute, National Institutes of Health under RFA # CA- 06-503 and through cooperative agreements with members of the Breast Cancer Family Registry (BCFR) and principal investigators. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the BCFR, nor does mention of trade names, commercial products or organizations imply endorsement by the US government or the BCFR. We wish to thank Teresa Selander, Nayana Weerasooriya and members of the Ontario Cancer Genetics Network for their contributions to the study.
Epidemiological study of BRCA1 and BRCA2 mutation carriers (EMBRACE).
Douglas F. Easton is the PI of the study. EMBRACE Collaborating Centers are Coordinating Centre, Cambridge: Susan Peock, Margaret Cook, Clare T. Oliver, Debra Frost, Radka Platte. North of Scotland Regional Genetics Service, Aberdeen: Zosia Miedzybrodzka, Helen Gregory. Northern Ireland Regional Genetics Service, Belfast: Patrick Morrison, Lisa Jeffers. West Midlands Regional Clinical Genetics Service, Birmingham: Trevor Cole, Kai-ren Ong, Jonathan Hoffman. South West Regional Genetics Service, Bristol: Alan Donaldson, Margaret James. East Anglian Regional Genetics Service, Cambridge: Joan Paterson, Sarah Downing, Amy Taylor. Medical Genetics Services for Wales, Cardiff: Alexandra Murray, Mark T. Rogers, Emma McCann. St James’s Hospital, Dublin & National Centre for Medical Genetics, Dublin: M. John Kennedy, David Barton. South East of Scotland Regional Genetics Service, Edinburgh: Mary Porteous, Sarah Drummond. Peninsula Clinical Genetics Service, Exeter: Carole Brewer, Emma Kivuva, Anne Searle, Selina Goodman, Kathryn Hill. West of Scotland Regional Genetics Service, Glasgow: Rosemarie Davidson, Victoria Murday, Nicola Bradshaw, Lesley Snadden, Mark Longmuir, Catherine Watt, Sarah Gibson, Eshika Haque, Ed Tobias, Alexis Duncan. South East Thames Regional Genetics Service, Guy’s Hospital London: Louise Izatt, Chris Jacobs, Caroline Langman, Anna Whaite. North West Thames Regional Genetics Service, Harrow: Huw Dorkins, Kashmir Randhawa. Leicestershire Clinical Genetics Service, Leicester: Julian Barwell, Nafisa Patel. Yorkshire Regional Genetics Service, Leeds: Julian Adlard, Carol Chu, Julie Miller. Merseyside & Cheshire Clinical Genetics Service, Liverpool: Ian Ellis, Catherine Houghton. Manchester Regional Genetics Service, Manchester: D Gareth Evans, Fiona Lalloo, Jane Taylor. North East Thames Regional Genetics Service, NE Thames, London: Lucy Side, Alison Male, Cheryl Berlin. Nottingham Centre for Medical Genetics, Nottingham: Jacqueline Eason, Rebecca Collier. Northern Clinical Genetics Service, Newcastle: Fiona Douglas, Oonagh Claber, Irene Jobson. Oxford Regional Genetics Service, Oxford: Lisa Walker, Diane McLeod, Dorothy Halliday, Sarah Durell, Barbara Stayner. The Institute of Cancer Research and Royal Marsden NHS Foundation Trust: Ros Eeles, Susan Shanley, Nazneen Rahman, Richard Houlston, Elizabeth Bancroft, Lucia D’Mello, Elizabeth Page, Audrey Ardern-Jones, Kelly Kohut, Jennifer Wiggins, Elena Castro, Anita Mitra, Lisa Robertson. North Trent Clinical Genetics Service, Sheffield: Jackie Cook, Oliver Quarrell, Cathryn Bardsley. South West Thames Regional Genetics Service, London: Shirley Hodgson, Glen Brice, Lizzie Winchester, Charlotte Eddy, Vishakha Tripathi, Virginia Attard. Wessex Clinical Genetics Service, Princess Anne Hospital, Southampton: Diana Eccles, Anneke Lucassen, Gillian Crawford, Donna McBride, Sarah Smalley. EMBRACE is supported by Cancer Research UK Grants C1287/A10118 and C1287/A11990. D. Gareth Evans and Fiona Lalloo are supported by an NIHR grant to the Biomedical Research Centre, Manchester. The Investigators at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust are supported by an NIHR grant to the Biomedical Research Centre at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust. Ros Eeles, Elizabeth Bancroft and Lucia D’Mello are also supported by Cancer Research UK Grant C5047/A8385.
CBCS: We thank Bent Ejlertsen, Mette K. Andersen, Anne-Marie Gerdes and Susanne Kjaergaard for clinical data. Moreover, we thank the NEYE foundation for financial support.
Conflict of interest
The authors report no conflicts of interest.
Author information
Authors and Affiliations
Consortia
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Pattern of linkage disequilibrium across BRCA1 in 216 Ashkenazi women with the founder 185delAG mutation. This is a portion of the plot generated by Haploview. The asterisk (*) marks the haplotype block used as the core to determine haplotype structure. Note: LD for women carrying the 5382insC mutation looks nearly identical (data not shown).
Supplementary Fig. 2 Recombination landscape across 2.1-Mb region surrounding BRCA2. LDHat with a block penalty of ten was used to estimate population recombination rates (4Nr, where N is the effective population size and r is the recombination fraction) in 372 Ashkenazi women. The Markov chain was updated 1 million times and estimates were sampled every 2000 iterations. Gray bar is the location of the BRCA2 gene.
Supplementary Fig. 3 The inbreeding coefficient (FIS) among 372 Ashkenazi women across chromosome 13. Positive values reflect a deficiency of heterozygosity compared to what is expected under Hardy–Weinberg equilibrium. Black lines represent the mean F IS ± 3 SD along chromosome 13. Black box marks the location of the 2.1 Mb BRCA2 region on chromosome 13.
Appendix A—msHOT command
Appendix A—msHOT command
/mshot 412 1 -r 1.0 2087508 -s 545 -v 30 46440 55764 92.72 94074 101467 172.87 116262 120708 84.46 158286 166715 60.34 188386 195853 69.08 203508 207586 27.50 408508 419950 23.61 425890 436582 14.86 531508 540447 22.26 661931 673793 120.99 770385 780441 29.47 815508 819627 29.47 824410 832053 71.03 896479 899758 92.66 907490 914508 66.72 927238 930508 5.91 951526 958508 33.76 967928 979794 50.26 996275 999508 107.56 1084508 1088508 28.62 1091504 1098508 126.14 1616508 1619571 340.72 1621508 1627508 15.09 1660329 1663531 623.28 1675309 1679508 35.42 1686508 1706508 5.02 1850331 1854508 65.25 1883508 1887508 237.76 1962482 1967508 38.82 2055476 2087508 53.91 -G 2355557.4 -eG 0.0000018 0.0000571 -eN 0.0000018 0.095.
Rights and permissions
About this article
Cite this article
Im, K.M., Kirchhoff, T., Wang, X. et al. Haplotype structure in Ashkenazi Jewish BRCA1 and BRCA2 mutation carriers. Hum Genet 130, 685–699 (2011). https://doi.org/10.1007/s00439-011-1003-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1003-z