Abstract
Changes in epigenetic marks such as DNA methylation and histone acetylation are associated with a broad range of disease traits, including cancer, asthma, metabolic disorders, and various reproductive conditions. It seems plausible that changes in epigenetic state may be induced by environmental exposures such as malnutrition, tobacco smoke, air pollutants, metals, organic chemicals, other sources of oxidative stress, and the microbiome, particularly if the exposure occurs during key periods of development. Thus, epigenetic changes could represent an important pathway by which environmental factors influence disease risks, both within individuals and across generations. We discuss some of the challenges in studying epigenetic mediation of pathogenesis and describe some unique opportunities for exploring these phenomena.
Similar content being viewed by others
Background
The field of epigenetics grew from attempts, beginning over 70 years ago, to understand mechanisms whereby multiple cellular phenotypes arise from a single genotype during the complex process of developmental morphogenesis termed epigenesis. The term “epigenetics” was initially reserved for mechanisms by which phenotypic state, as determined by differential gene expression, could be stably retained through cell division by non-genetic factors. Various mechanisms have been proposed to have the potential to encode this phenotypic information; these include enzymatic methylation of cytosine bases (DNA methylation), post-translational modification of tail domains of histone proteins (histone modifications) and associated nucleosome positioning or chromatin remodeling, non-coding RNAs, and transcription factor regulatory networks (Ptashne 2007). Epigenetic marks established by each of these processes are often shared within a cell lineage; however, whether all persisting epigenetic marks satisfy requirements for stable transmission through cell division or some are merely reestablished from other information following mitosis remains a vigorously debated question.
The term epigenetics has more recently been used in the scientific literature to describe various unspecified non-genetic mechanisms influencing phenotype. This broader usage emerged from mouse studies addressing transgenerational nutritional effects on phenotype, as well as human studies of phenotypic differences between monozygotic twins. In the popular press “epigenetics” has become almost synonymous with nutritional and environmental influences on gene expression. Thus, while “epigenetics” initially referred to largely self-contained developmental processes, it has come to describe environmental influences on phenotypic readout of genotypes. This semantic evolution has caused confusion and controversy regarding the meaning of “epigenetics” at a time of intensified interest in the possible role of epigenetic mechanisms in disease. In this review we define as epigenetic processes those that stably affect gene expression through mechanisms not involving the primary nucleotide sequence, and epigenetic state as the configuration of chromatin and DNA marks utilized by these processes. By contrast, genetic state is widely understood to refer to the primary nucleotide sequence itself, while genetic processes maintain or change nucleotide sequence.
Epidemiologic research addressing epigenetic mechanisms as mediators of environmental exposures on disease risk is constrained by important ethical considerations. These often preclude both experimental exposure to candidate environmental causes, and invasive collection of cell types of greatest developmental and functional relevance to disease processes. Inquiry has therefore progressed largely by integrating information about biological mechanisms obtained in model systems with observational data provided by humans. To address the current state and future promise of this research, we undertook this review with two goals: to illustrate the potential of epigenetic processes to mediate exposure–phenotype relationships and to discuss study design and statistical analysis methods needed to investigate such mechanisms in relation to origins of human disease. We begin by discussing genetic, developmental, and environmental determinants of epigenetic state in human and model systems, then describe some of the diverse data implicating epigenetic mechanisms in various human diseases, both within individuals and across generations. We conclude by discussing technical challenges, suggesting promising opportunities for epidemiologic research in environmental epigenetics, and offering some thoughts about translational significance and future directions of this field.
Determinants of epigenetic state
Epigenetic mechanisms work in concert to influence the potential for gene expression at myriad locations throughout the genome. The resulting epigenetic state of the genome, termed epigenome, varies by cell type. Considering the tremendous diversity of epigenetic marks, which include dozens of different post-translational histone modifications and more than 50 million sites of potential DNA methylation in a diploid human genome (and thus >250M possible epigenotypes!), it seems that no two human cells would have identical epigenomes. Indeed, within each individual there are many epigenomes, and these change over time as a consequence of both normal developmental and pathological processes, as well as environmental exposures and random drift. Despite this potential for considerable variability of epigenetic patterns within and between individuals, there can also be remarkable consistency. In a study of 11 tissues in 6 autopsies, DNA methylation patterns in a highly selected set of loci were found to be highly conserved, with intraclass correlations of 0.85 across tissues within individuals and 0.83 across individuals within tissues (Byun et al. 2009). The authors interpreted these patterns as revealing different sets of person-specific and tissue-specific differentially methylated genes, anticipating subsequently observed differential genetic and acquired determination (Waterland et al. 2010).
DNA methylation has been the epigenetic mark most extensively measured in epidemiologic research for numerous reasons. It is of fundamental biological interest owing to its unambiguously stable transmission during cell division. It also has practical advantages: as a chemically stable covalent change to the DNA itself, DNA methylation is the only epigenetic mark that survives the DNA extraction and purification that is routine in molecular sample processing, and it can endure decades of archival sample storage (Kristensen et al. 2009).
Genetic influences
Nucleotide sequence is a primary determinant of epigenetic state, clearly evident from the distribution of epigenetic marks across the genome, determined in part by direct effects of G:C content and CpG (cytosine-phosphate-guanine dinucleotide) density (Tanay et al. 2007; Thomson et al. 2010). Additional genetic influences include proximity to repetitive elements such as Alu and LINE1 (Estecio et al. 2010), nuclear architecture (Berman et al. 2011), and binding sequences for transacting proteins (Bell et al. 2011b; Weth and Renkawitz 2011). Motif searches and screening strategies have identified sequence elements that predispose to particular epigenetic states (Feltus et al. 2006; Ideraabdullah et al. 2011; Keshet et al. 2006; Lienert et al. 2011).
Several lines of evidence indicate that genetic polymorphisms can affect epigenetic state. Greater differences were observed between dizygotic co-twins than between monozygotic co-twins in two forms of epigenetic state: skewed patterns of X-inactivation, and DNA methylation at differentially methylated regions of the imprinted IGF2/AH19 locus (Wong et al. 2011; Heijmans et al. 2007; Ollikainen et al. 2010). Extensive DNA methylation analyses in a multigenerational family revealed that epiallelic similarity was greater among first-degree relatives than among more distantly related family members. In the same study, analyses addressing both genetic variation and DNA methylation identified widespread occurrence of allele-specific DNA methylation (ASM) that was associated with polymorphic nucleotides located near the DNA methylation site, but not parent of origin. Authors of this report concluded that the majority of such ASM events depend on cis-acting DNA sequence (Gertz et al. 2011). Such ASM events have yet to be characterized in large population-based studies, but more modest studies addressing heterozygous non-imprinted loci have identified widespread ASMs associated with nearby genotypic polymorphisms in DNA from multiple tissue types (Kerkel et al. 2008; Tycko 2010; Schalkwyk et al. 2010), as well as allele-specific chromatin structure and transcription factor binding in lymphoblastoid cell DNA (McDaniell et al. 2010, reviewed in Birney et al. 2010). Presumed transgenerational inheritance of epigenetic changes (“epimutations”) in the MLH1 (Suter et al. 2004) and MSH2 (Chan et al. 2006) mismatch repair genes, both associated with colorectal cancer, were also traced to germline genetic variation. In the case of the MSH2 epimutation, deletion of a gene immediately upstream of the MSH2 gene causes transcription to run through the MSH2 promoter, causing somatic hypermethylation and gene silencing (Ligtenberg et al. 2009). The MLH1 epimutation was found to be caused by a polymorphism in the 5′ UTR of the MLH1 gene, reducing transcriptional activity, and predisposing to aberrant somatic DNA methylation in each generation (Hitchins et al. 2011).
Developmental programming of the epigenome
In successful mammalian reproduction, the single-cell zygote gives rise to an organism with hundreds of cell types. These diverse cellular phenotypes arise from the same shared genomic sequence by control of the subset of genes expressed in each cell type. Cellular differentiation is tightly linked to extensive erasure and establishment of lineage-specific epigenetic marks, a process termed epigenetic reprogramming. Relatively detailed descriptions of DNA methylation in developing tissues have been carried out in the mouse, which serves as a model of epigenetic reprogramming in mammalian development (Trasler 2009).
At fertilization, reprogramming begins with extensive erasure of methyl marks in DNA of the paternal (sperm-derived) DNA, followed by more general loss of methyl marks in the zygote and embryo during cleavage divisions, while sparing parent-of-origin specific imprints. By the blastocyst stage, de novo DNA methylation distinguishes inner cell mass cells (from which embryonic lineages arise to create fetal structures) from relatively hypomethylated trophectoderm cells (from which extra-embryonic lineages arise to create transient structures, including placenta) (Fig. 1).
Reprogramming of DNA methylation in the zygote, early embryo, and primordial germ cells. Thickness of the outer arrows indicates levels of DNA methylation. Red maternal genome, blue paternal genome, black diploid genome. Embryonic lineages arise from cells of the inner cell mass (ICM), the placenta and extraembryonic membranes from trophectoderm cells, and the germ cell lineage from primordial germ cells following their determination from proximal epiblast. Inner circles indicate developmental stages when key elements of epigenetic programming are thought to occur (Adapted from Feng et al. 2010)
Germ cell lineage specification begins in cells of the proximal epiblast, and involves a second extensive erasure of DNA methylation that removes parental imprint marks (Fig. 1). Thereafter, the germ line develops in a sexually dimorphic fashion. New DNA methyl marks are established over many stages, extending through sexual maturity in accordance with the sex of the developing individual. At this time, the sex-specific imprint marks that govern parent-of-origin specific expression of imprinted genes in the subsequent generation are established (Faulk and Dolinoy 2011).
Developmental reprogramming can result in dramatic epigenetic differences between the two alleles. The association between mono-allelic gene expression and DNA methylation has long been recognized, both in the context of X-inactivation in females (Boggs et al. 2002; Sharp et al. 2011) and in parent-of-origin determined genomic imprinting (Ferguson-Smith 2011), but now also in the mono-allelic expression of non-imprinted autosomal loci (Harris et al. 2010; Tarutani and Takayama 2011).
Further resetting of epigenetic marks accompanies differentiation of many specialized cell types of the body as well as placenta and other transient structures during pregnancy, and subsequent development of body structures during various postnatal stages of development. Chromatin states that arise during development can affect the propensity to subsequent epigenetic change. An example of this is the predisposition of polycomb-repressive complex occupied genes in stem cells to the acquisition of DNA methylation abnormalities in aging and cancer (Ohm et al. 2007; Schlesinger et al. 2007; Teschendorff et al. 2010; Widschwendter et al. 2007).
Environmental influences
Multiple differences in gene expression, presumably reflecting intrauterine epigenetic differences, have been identified in several tissues from newborn identical twins (Gordon et al. 2011). The global methylation pattern of individuals changes with increasing age (Bjornsson et al. 2008), as does the difference in global methylation between MZ twin pairs (Fraga et al. 2005). Genetically identical MZ twins show some epigenetic discordance at birth, as indicated by gene expression discordance (Gordon et al. 2011). Even over the first decade of life (Wong et al. 2010), and as aging adults (Talens et al. 2010), MZ twins acquire additional differences in epigenetic state, which may partly reflect different exposure histories, as would be expected if environmental exposures influence epigenetic state. However, stochastic drift in epigenetic state and related consequences such as mono-allelic expression described in the previous section is likely responsible for much of the observed divergence. Therefore, other forms of data (discussed below) are needed to determine what type of exposures may influence epigenetic state and the extent of resulting changes.
Experimental studies
The most direct evidence suggesting that ambient exposures may influence epigenetic state is experimental. In vitro studies have demonstrated associations of DNA methylation with various metals (Dolinoy et al. 2007b; Wright and Baccarelli 2007). In the in vivo setting, prenatal protein restriction is associated with hypomethylation of the glucocorticoid receptor (GR) and PPARα gene promoter regions in rat liver (Lillycrop et al. 2005), changes that were prevented by folic acid supplementation (Lillycrop et al. 2005) and which were transmitted to the F2 generation (Burdge et al. 2007). Plagemann et al. (2009) found hypermethylation in the promoter of the anorexigenic gene for proopiomelanocortin in rats overfed as neonates. Whether this change could be transmitted to offspring was not assessed. DNA from sperm of mice exposed to steel plant air was found to be persistently hypermethylated long after exposure ended (Yauk et al. 2008). Additionally, maternal nurturing behavior has been shown to modify methylation at individual CpG sites in the ngf1a binding region of the GR gene in the hippocampus of the offspring (Weaver et al. 2004), an epigenetic modification that persisted both into adulthood to modify response to stress, and into the F2 generation.
Human studies
Christensen and Marsit (2011) and Terry et al. (2011) have provided comprehensive reviews of environmental influences on epigenetic state in humans. Here we note exposure periods of particular interest and several examples of environmental exposures reportedly associated with epigenetic state of specific human cell types.
The epigenome may be most vulnerable to environmental insults during periods of extensive epigenetic reprogramming, which may in theory be disrupted by exposures that interfere with any process that governs reprogramming. Periods of particular vulnerability may therefore include the early stages of embryonic development mentioned above. Childhood is also proposed as a period of vulnerability, especially in the germline of females, since oocytes remain in a haploid de-methylated state until puberty, so environmental insults may potentially disturb the epigenetic state of the oocyte for many years (Faulk and Dolinoy 2011), with potential implications for both fertility and initial epigenetic state of offspring of an exposed female. Somatic changes to DNA methylation may also result from environmental exposures in adults, as have been observed in aging and disease processes such as cancer described in the next section.
Energy and nutrient intake
Significant epigenetic changes in the IGF2 gene have been documented in those prenatally exposed to severe caloric restriction during the Dutch hunger winter of World War II (Heijmans et al. 2008). Hughes et al. (2009) additionally found that those most likely to be exposed to this famine during adolescence or young adulthood had a significantly decreased risk of developing colorectal cancers (CRC) characterized by the CpG island methylator phenotype (CIMP), suggesting a role for early life exposures in CIMP-specific CRC pathogenesis.
Folates are the major source of the methyl groups used for DNA and histone methylation. One study of folates and other one-carbon nutrients reported a differential effect of folate on the risk of the CIMP CRC subset compared to the non-CIMP subset (Van Guelpen et al. 2010), while two other studies did not (Slattery et al. 2006; van den Donk et al. 2007). Most studies of the microsatellite instability high subset, characterized by hypermethylation of the MLH1 gene promoter region and CIMP (Weisenberger et al. 2006), have yielded similarly negative results (Eaton et al. 2005; Schernhammer et al. 2008; Slattery et al. 2001; Wark et al. 2005). On the other hand, in some studies the association between intake of alcohol (which degrades folates) and CRC risk has been reported to be greater in MSI-H and CIMP tumors (Diergaarde et al. 2003; Eaton et al. 2005; Slattery et al. 2001).
Micro-RNAs (miRNAs) are very short non-coding RNA molecules that can downregulate protein-coding genes by destabilizing mRNAs or blocking translation. The possibility that exogenous microRNA consumed in food may epigenetically regulate gene expression has emerged from recent studies demonstrating the presence of plant-derived miRNAs in sera of humans and other mammals (Zhang et al. 2012). One of these plant microRNAs, MIR168a, which was demonstrated to be only of plant origin in control mice, binds coding sequence of the mammalian LDLRAP1 gene in vitro. Functional consequences in mammalian systems were demonstrated experimentally, as MIR168a administered in vitro and during in vivo feeding studies decreased expression of the protein product of LDLRAP1. This line of research suggests novel epigenetic mechanisms whereby diet may modify risk of human disease.
Air pollution
Emerging evidence suggests that air pollutants can influence epigenetic changes, including DNA methylation as well as up- or down-regulation of miRNAs (Jardim 2011). In human epidemiologic studies, PM2.5 and PM10 exposures are associated with hypomethylation of Alu and/or LINE1 elements in leukocytes and buccal cells (Baccarelli et al. 2009; Bollati and Baccarelli 2010; Madrigano et al. 2011; Salam et al. 2012; Tarantini et al. 2009), as well as altered DNA methylation in NOS2A, a gene involved in production of nitric oxide (Salam et al. 2012; Tarantini et al. 2009). Living in highly polluted cities (high PM and ozone) is also associated with hypermethylation of FOXP3 in regulatory T cells (Nadeau et al. 2010), while neonates who were prenatally exposed to polyaromatic hydrocarbon (PAH) had hypermethylated ACSL3 in DNA of umbilical cord white blood cells (Perera et al. 2009); notably, both of these genes are involved in asthma pathogenesis. PAHs are also associated with hypermethylation of LINE1 and Alu (Pavanello et al. 2009; Perera et al. 2009). More limited evidence is emerging to suggest that air pollution is associated with changes in miRNA expression (Bollati et al. 2010; Jardim 2011), and adverse effects of air pollution constituents may be modified by variant alleles of genes involved in miRNA processing (Wilker et al. 2010).
Tobacco smoke
Fetal exposure to maternal smoking during pregnancy (PTS) is associated with reduced methylation of several repeated sequences, including Sat2 (Flom et al. 2011), Alu, and LINE1 among children with the GSTM1 null genotype (Breton et al. 2009). PTS exposure is also associated with increased DNA methylation in specific genes, such as AXL and PTPRO (Breton et al. 2009, 2011b) and IGF2 (Murphy et al. 2011). In adult lung cancer patients, quantity and duration of active smoking as well as second-hand smoke is associated with increased DNA methylation of p16 (Kim et al. 2001; Scesnaite et al. 2012), MGMT, and DAPK (Russo et al. 2005). Tobacco smoke is also associated with tumor cell DNA methylation changes in esophageal squamous cell carcinoma (Huang et al. 2011), significantly higher frequencies of abnormal DNA hypermethylation in prostate (Enokida et al. 2006) and gastric cancers tumor cells (Nan et al. 2005) and with a higher risk of CIMP + colorectal tumors (Limsui et al. 2010; Samowitz et al. 2006). Lastly, the F2RL3 gene is hypomethylated in smokers and may mediate the detrimental impact of smoking on cardiovascular mortality, since hypomethylated F2RL3 was found to be strongly associated with cardiovascular mortality among patients with stable coronary heart disease (Breitling et al. 2012).
Oxidative stress
Reactive oxygen species (ROS) are involved in numerous cellular processes including cellular redox alterations, immune response, signaling pathways, chromatin remodeling and gene expression (Sundar et al. 2010). ROS have the potential to influence epigenetic mechanisms (Baccarelli and Bollati 2009), and have been shown to inhibit binding of methyl-CpG binding protein 2, a critical epigenetic regulator that recruits cytosine methyl transferases and histone deacetylases to DNA (Valinluck et al. 2004). Numerous environmental exposures, including constituents of air pollution and tobacco smoke, can generate ROS and thus may potentially alter epigenetic processes through oxidative stress mechanisms.
Metals
Prenatal lead exposure is associated with decreased methylation of LINE1 and Alu in cord blood (Pilsner et al. 2009), and a similar pattern of LINE1 methylation was reported in an elderly cohort (Wright et al. 2010). Studies in humans have shown that arsenic exposure is associated with either global hypermethylation or hypomethylation in peripheral blood mononuclear cells (PBMCs) depending on dose (Majumdar et al. 2010), as well as DNA hypermethylation of several genes, including CDKN2A (Chanda et al. 2006), RASSF1A and PRSS3 (Marsit et al. 2006). Exposure to airborne particulates rich in lead, cadmium and chromium are associated with miRNA expression in peripheral blood (Bollati et al. 2010) and airborne levels of nickel and arsenic are positively correlated with both histone 3-lysine4 trimethylation (H3K4me3) and histone 3-lysine9 acetylation (H3K9ac) in blood leukocytes (Cantone et al. 2011). Occupational exposure to nickel is associated with increased H3K4me3 and decreased H3K9me2 in PBMCs (Arita et al. 2011). Lastly, cadmium can induce overexpression of the DNA methyltransferase genes DNMT1 and DNMT3a in human embryo lung fibroblasts, and is associated with hypermethylation and silencing of the MSH2, ERCC1, XRCC1 and OGG1 genes in human bronchial epithelial cells (Jiang et al. 2008; Zhou et al. 2011).
Organic chemicals
Gas-station attendants and police officers occupationally exposed to low levels of benzene were found to have significantly lower LINE1 and Alu methylation, hypermethylation of p15, and hypomethylation of MAGE-1 in blood (Bollati and Baccarelli 2010; Bollati et al. 2007).
Genetic × epigenetic × environmental interactions
Most work investigating effects of environmental factors on epigenetic state has not considered the potential for genetic susceptibility to modify these associations. However, Salam et al. (2012) recently investigated contributions of both genetic and epigenetic variation in air pollution-mediated levels of fractional exhaled nitric oxide (feNO). Measurement of feNO provides an in vivo summary assessment of inducible nitric oxide synthase (iNOS) activity as well as airway inflammation. These investigators found interrelated effects of exposure to the air pollution constituents PM2.5, NOS2A promoter haplotypes, and methylation of the iNOS encoding gene NOS2A and NOS2 promotor haplotypes on feNO level. These observations illustrate not only the feasibility of assessing interactions between epigenetic, genetic, and environmental factors, but also the importance of doing so in order to delineate complex biological relationships and identify susceptible subpopulations.
Epigenetic effects in human disease
Conditions associated with improper parental contributions of imprinted genes are currently the clearest examples of human diseases related to epigenetic state. Even before genomic imprinting was described, experiments in which pronuclei were transplanted into enucleated eggs demonstrated that both maternal and paternal chromosomal contributions are required for normal development. Control conceptuses receiving one set (haploid genome) of maternal (egg-derived) and one set of paternal (sperm-derived) chromosomes could develop normally. However, abnormal development and early demise occurred in all conceptuses receiving either two maternal sets or two paternal sets of chromosomes (McGrath and Solter 1984), which developed into tissues with histologic features of dermoid cysts and hydatiform moles, respectively.
Model imprinting disorders such as Beckwith–Wiedemann, Angelman, Russell–Silver, and Prader–Willi syndromes are human conditions that can be caused by aberrant epigenetic state (Ferguson-Smith 2011). The specific features, early onset, and rarity of these disorders facilitated recognition of their relation to improper parental contributions of imprinted loci (e.g. two maternal or two paternal copies, rather than one maternal and on paternal copy), which can arise from uniparental disomy, more local epigenomic abnormalities (Tierling et al. 2011), or widely distributed changes caused by genetic defects in trans-acting epigenetic regulators (Begemann et al. 2011).
The role of epigenetics in most other conditions remains far less clear. However, since Barker proposed that there is sometimes a fetal basis for adult disease (Barker et al. 1993), epidemiologic research has implicated numerous environmental exposures during prenatal and early postnatal development as influencing risk of adult cardiovascular disease, obesity, type 2 diabetes, and other chronic conditions (Taylor and Poston 2007). Epigenetic processes have been proposed to explain lengthy time periods between exposure and disease onset: if exposures during early periods of epigenetic vulnerability influence epigenetic state, resulting epigenetic marks may be stably maintained in cell lineages to influence disease susceptibility years later. Waterland and Jirtle (2004) and Lillycrop (2011) recently discussed experimental data addressing the plausibility of such mechanisms. Human research described here provides correlative data. Associations between exposure history and epigenetic state are described in the previous section, while associations between epigenetic state and disease risk are outlined below, with a few studies reporting associations over the exposure—epigenetic state—disease continuum.
Reproductive conditions
The possibility that epigenetic state can influence reproductive success was recognized once research in model organisms revealed that dramatic epigenetic reprogramming occurs during gametogenesis, fertilization, and development of the zygote, embryo and placenta (Feng et al. 2010). Ethical considerations have limited studies that directly measure epigenetic state to a small subset of relevant cell types and developmental stages. Here we review selected data addressing the hypothesis that epigenetic phenomena may influence risk of infertility, disorders of placental development and function, and birth weight.
Human infertility research has assessed DNA methylation profiles in DNA isolated from spermatozoa, fully differentiated male germline cells that are amenable to noninvasive collection. Although neither measures of DNA methylation nor outcomes have been standardized across published studies, most report variation in epigenetic state of sperm DNA to be associated with undesirable outcomes (Carrell and Hammoud 2010). Early studies employing methylated cytosine immunostain—a genome-wide measure without sequence specificity—revealed little difference between sperm DNA methylation and sperm quality (Benchaib et al. 2003) or fertilization rate and embryo quality following in vitro fertilization (IVF) (Benchaib et al. 2005). However, all subsequent studies assaying specific sequence elements (Pacheco et al. 2011, reviewed in Cortessis et al. 2011; van Montfoort et al. 2012) reported abnormal methylation of one or more imprinted genes in DNA from poor quality sperm, as predicted if sperm production defects involve deregulated imprinting (Filipponi and Feil 2009). Three studies assaying repeated elements reported no association between sperm quality and methylation of Alu (Kobayashi et al. 2007) or LINE1 (Houshdaran et al. 2007; Marques et al. 2008), while one reported elevated Alu methylation in sperm used in assisted reproductive technologies (ART) leading to pregnancy and live birth (El Hajj et al. 2011) and another that poor sperm quality was associated with hypermethylation of the repetitive element SAT2CHRM1 (Houshdaran et al. 2007). Studies interrogating CpG islands not associated with recognized imprints reported methylation at numerous sites to be associated with sperm quality, implicating widespread disruption of DNA methylation in abnormal spermatogenesis (Houshdaran et al. 2007; Pacheco et al. 2011). This avenue of research is expanding to address additional epigenetic marks of specific relevance to the male germline (Carrell and Hammoud 2010; van Montfoort et al. 2012).
Early stages of germ cell development are reportedly identical in both sexes, so early disruptions of epigenetic state could in theory impair fertility of both males and females. However, direct epigenetic study of female gametes is not regarded as practicable, because very few oocytes are produced, and oocyte retrieval procedures may alter epigenetic state (Fortier et al. 2008; Li et al. 2011; Sato et al. 2007).
Epigenetic state has also been implicated in infertility by a distinct form of data, excess occurrence of model imprinting disorders among children conceived by ART (van Montfoort et al. 2012). A recent meta-analysis found elevated occurrence of imprinting disorders of both maternal origin (Angelman syndrome, Beckwith–Wiedemann syndrome) and paternal origin (Prader–Willi syndrome) (Cortessis 2009). Because only sub-fertile individuals use ART, these results could indicate a tendency for sub-fertile adults to produce gametes with inappropriately set imprint marks. However, whether underlying molecular defects of these children are epigenetic or genetic was not reported, and even if they were epigenetic, alternate explanations are credible. Couples undergoing ART are dissimilar to the general population with respect to age and income and thus numerous unmeasured factors that could be postulated to influence epigenetic state. Moreover, ART procedures bypass various functions that accompany unassisted conception. These include the need for spermatozoa to traverse the female reproductive tract and compete to penetrate the oocyte, which may serve to select for fertilization sperm with normal epigenetic state. Finally, effects of ART themselves have been postulated, since some imprinting disorders share overgrowth features of the veterinary condition large offspring syndrome, which appears to be inducible by embryo culture (Sinclair et al. 2000). However, data on epigenetic state of human oocytes and embryos are extremely limited (van Montfoort et al. 2012).
Fetal survival, development and growth depend upon the placenta. Arising from the relatively hypomethylated TE, cells of the developing placenta undergo extensive de novo DNA methylation (Serman et al. 2007). Epigenetic regulation of placental function changes during gestation (reviewed in Nelissen et al. 2011), accompanied by a dynamic process of loss of imprinting postulated to have an important role in placental maturation and function (Pozharny et al. 2010). Unidentified placental functions are postulated to rely on imprinting, because imprinted genes are abundantly expressed in the placenta, compared with other organs, and imprinting arose during mammalian evolution (Nelissen et al. 2011). Disturbed placental function may cause intra-uterine growth restriction (IUGR), a predictor of poor health in the neonatal period and childhood, which may also predispose to diseases of adulthood. Two small studies examining mRNA transcript levels in placentas of IUGR and normal birthweight infants reported differential expression of some placentally imprinted genes (Diplas et al. 2009; McMinn et al. 2006); a third reported lower IGF2 and loss of IGF2 imprints (Koukoura et al. 2011a) as well as higher H19 transcript levels and increased H19 expression (Koukoura et al. 2011b) in growth-restricted placentas. Differential levels of transcripts encoded by individual imprinted loci were also reported in IUGR placentas (Apostolidou et al. 2007) and small for gestational age infants (Guo et al. 2008).
Infants conceived by ART have been shown consistently to have lower birthweight than other infants (Andersen et al. 2008). A recent meta-analysis revealed that in published studies of singleton births controlled for effects of maternal age, low birthweight was more common among those conceived by IVF (McDonald et al. 2009). In the same review, IVF conception was also associated with premature birth, which is highly associated with low birthweight. Similar analyses indicate that low birthweight and preterm birth are also more common among twins conceived by IVF compared with naturally conceived twins (McDonald et al. 2010). Addressing ART procedures more generally, recently published surveillance data on over 16,000 births revealed elevated occurrence of low birthweight, small-for-gestational age, and preterm birth among singleton but not twin infants conceived following ART (D’Angelo et al. 2011).
It is presently not clear whether low birthweight following ART reflects epigenetic state of gametes inherited from sub-fertile parents, effects of ART, or other unrecognized factors. Additional insight may accrue from future research addressing possible reasons that ART conception is associated with model imprinting disorders and low birthweight, as well as recently initiated investigation of any relationship between epigenetic state and these outcomes in naturally conceived neonates (Lee et al. 2012).
Asthma
Emerging evidence suggests that the epigenetic state may contribute to asthma pathogenesis. Asthma is an IgE-mediated type I hypersensitivity reaction involving skewed programming of naïve CD4+ T lymphocytes toward a Th2 lineage (Durham et al. 2011). Shift toward a Th2 phenotype may result from chromatin remodeling brought about by multiple, coordinated epigenetic changes on genes regulating Th differentiation (Ho 2010). These include loss of DNA methylation and gain of H3K9ac and H3L4me2 in IL-4 and IFN-γ, genes that encode key cytokines involved in T cell lineage development (Fields et al. 2002; Jones and Chen 2006; Kwon et al. 2008; White et al. 2002). DNA methylation of the Foxp3, ACSL3, and ARG2 loci are associated with impaired regulatory T-cell function (Nadeau et al. 2010), increased asthma morbidity (Nadeau et al. 2010; Perera et al. 2009), and exhaled nitric oxide production, respectively (Breton et al. 2011a). Histone deacetylases (HDACs) have also been shown to maintain pre-established Th1-like and Th2-like immunity in human T cells; inhibition of HDAC can shift the Th1:Th2 ratio, skewing response toward Th2-like phenotype (Su et al. 2008). Additional evidence implicates miRNAs in asthma pathogenesis (Pauley and Chan 2008). MiR-126 expression-suppressed asthmatic phenotype (Mattes et al. 2009) and differentially expressed miRNAs including miR-21, -103, -155, -146a, and -204 were identified in response to innate and adaptive immune stimuli (Liu et al. 2009; Nana-Sinkam et al. 2009).
Metabolic diseases
Functional epigenetic changes have been observed in obesity (Campion et al. 2009), diabetes (Reddy and Natarajan 2011), cardiovascular disease (Wierda et al. 2010), and the constellation of abnormalities known as metabolic syndrome (Bruce and Hanson 2010). For example, adverse vascular effects of hyperglycemia can continue for several years following sufficient glycemic control, a phenomenon called metabolic memory (Siebel et al. 2010), and there is evidence from animal studies that this effect is associated with epigenetic changes in the promoter of the NFκβ subunit of the p65 gene in aortic endothelial cells (El-Osta et al. 2008) and increased acetylation at H3K9/K14 in a number of genes involved in endothelial dysfunction and chronic inflammation (Pirola et al. 2011). In women, promoter-region methylation of GNSAS and INSIGF in leucocytes was reportedly associated with an increased incidence of myocardial infarction (Talens et al. 2011), suggesting that epigenetic modification may be clinically relevant. Although various histone modifications have been demonstrated in vitro in relation to complications of hyperglycemia (Wierda et al. 2010), permanence of individual changes has not been established.
Neurological disorders
Historically, the field of epigenetics has focused on elucidating mechanisms for maintaining DNA methylation in dividing cells. However, recent work has discovered dynamic DNA methylation changes in non-dividing cells including neurons (reviewed in Mill et al. 2008), motivating studies of effects of environmental exposures on epigenetic marks in relation to neurological disorders.
Alzheimer’s disease, schizophrenia, and autism spectrum disorders show a variety of epigenetic anomalies (Akbarian 2010; Mill et al. 2008; Nguyen et al. 2010b; Schroeder et al. 2011). An epigenetic mechanism has been proposed to explain the association between famine during the prenatal period and schizophrenia risk (Lumey et al. 2011), as well as associations between expression of imprinted genes and both autism and schizophrenia (Badcock and Crespi 2008). Recent studies, moreover, report associations of both conditions with DNA methylation in additional, non-imprinted genes (Dempster et al. 2011; Mill et al. 2008; Nagarajan et al. 2006; Schroeder et al. 2011).
Cancer
Abnormal DNA methylation, histone code modifications and miRNA changes have all been demonstrated in human cancer cells as reviewed elsewhere (Kanwal and Gupta 2011; Lovat et al. 2011; Sharma et al. 2010). In malignant tumors, gradual genome-wide DNA hypomethylation is observed, concurrent with hypermethylation at normally unmethylated CpG islands (Baylin and Jones 2011; Feinberg 2007). Although a direct association between hyper- and hypo-methylation was not initially recognized (Bariol et al. 2003; Iacopetta et al. 2007), recent work suggests that the two phenomena may well be linked, and are confined to similar compartments of the genome (Berman et al. 2012). Although the targeting of abnormal hypermethylation in gene promoter regions is not completely understood, its association with occupancy by polycomb complex proteins (Ohm et al. 2007; Schlesinger et al. 2007; Widschwendter et al. 2007), which transiently repress genes involved in differentiation, suggests that such methylation may be associated with either de-differentiation or a failure of tissue stem cells to fully differentiate.
Although most cancers show aberrant methylation of some gene promoter regions, a subset of tumors is characterized by CIMP, a phenotype involving concurrent methylation of multiple gene promoter regions (Toyota et al. 1999; Weisenberger et al. 2006). This phenotype is best described in colorectal cancer, for which a carefully vetted set of markers (Weisenberger et al. 2006) allows epidemiologic studies of risk factor differences between CIMP and non-CIMP tumors (reviewed in Curtin et al. 2011). The association between smoking and colorectal cancer was reported to be limited to those with the CIMP phenotype (Limsui et al. 2010; Samowitz et al. 2006). Additionally, integration of the hepatitis B virus (HBV) into hepatic cell DNA has been associated with increased DNA hypermethylation in hepatocytes (Herceg and Paliwal 2011) or alternatively simply reflect a consequence of increased cell turnover. Chronic inflammation may also modify epigenetic marks on both histones and DNA (Bayarsaihan 2011). Other environmental factors have also been associated with aberrant DNA methylation in some human studies (reviewed in Lim and Song 2012) but further study of additional populations is needed.
The role of folates and other one-carbon nutrients, which are critical in the provision of methyl groups, has been assessed in many epidemiologic studies of abnormalities in methylation in CRC, with largely disappointing results (Curtin et al. 2011). However, folates and polymorphisms in genes of the folate pathway have been shown to modify global DNA methylation in circulating lymphocytes (Lim and Song 2012), although the relationship between such changes and changes in target tissues is not known. Additionally, the risk of various childhood malignancies may be significantly decreased in children exposed in utero to higher folate and other one-carbon nutrients, mainly via multivitamin use (reviewed in Ciappio et al. 2011). Although a direct correlation between maternal one-carbon nutrient levels and aberrant DNA methylation in either childhood or adult tumors has not been demonstrated, these studies provide a basis for future research into the relationship between prenatal exposure to methylation-associated nutrients and the abnormal methylation events so common in carcinogenesis.
Transgenerational phenomena
In theory, epigenetic marks in somatic cell lineages could be transmitted to daughter cells over the lifespan of an exposed individual, while marks established in germ cell lineage members could be transmitted to subsequent generations. Several examples of apparent transmission of an environmental insult across generations are described below. However, whether epigenetic mechanisms mediate these phenomena remains highly controversial (Bollati and Baccarelli 2010). Convincing evidence that an epigenetic mechanism is responsible for transgenerational effects requires determination of the phenotype in at least three generations following exposure. Taking as F0 the exposed pregnant mother, observations of F1 through F3 generations are required to exclude the possibility of direct effects of the exposure both on somatic cells of the F1 generation or on germ cells progenitors from which the F2 generation will arise, since both are present during the exposed pregnancy (Perera and Herbstman 2011). It is also possible that selection of pregnancies for viability could influence the distribution of genes that influence epigenetic marks.
Experimental animals
The best-documented example of transgenerational specification of a phenotype through DNA methylation is the determination of coat color and other traits (obesity, diabetes, and tumors) in agouti mice. Dams carrying the meta-stable Avy epiallele (who themselves display the agouti coat pattern) are more likely to produce agouti offspring, while dams of the same strain without the Avy epiallele (who have yellow coats) are more likely to produce yellow offspring. A continuous gradient in coat color is related to the penetrance of the Avy epiallele (Morgan et al. 1999; Waterland and Jirtle 2003) determined in part by methyl donors in the dam’s diet. The phytoestrogen genistein can also change the epigenetic state of this allele for several subsequent generations (Cropley et al. 2006; Dolinoy et al. 2007a; Lillycrop et al. 2005). While these observations convincingly argue for epigenetic determination of the agouti phenotype, the meta-stable Avy epiallele arose from insertion of a retrotransposon rather than from conventional environmental exposure.
In a separate model, experimental exposure of pregnant rats to endocrine-disrupting agents (vinclozolin, bisphenol A) produced changes in male infertility, body weight, and cancer risk in up to four subsequent generations (Anway et al. 2005; Anway and Skinner 2008). However, whether specific epigenetic marks determine this phenotype across these generations has not been established.
Human studies
Famine
Pembrey et al. (2006) described an association between paternal grandfather’s exposure to a famine in northern Sweden and mortality in their grandsons, whereas paternal grandmother’s food supply was associated instead with mortality in their granddaughters. Lumey (1992) reported effects on birth weight up to two generations following the Dutch famine in World War II.
Tobacco smoke
The Southern California Children’s Health Study has demonstrated an effect of grandmaternal smoking while pregnant on the asthma risk of their grandchildren, after controlling for maternal smoking and environmental tobacco smoke exposure (Li et al. 2005). Pembrey et al. (2006) also described effects of pre-adolescent paternal smoking on BMI in sons but not daughters. Although any of these phenomena could be due to epigenetic mechanisms, mutation to the DNA itself would be a plausible alternative, as has been shown for mainstream tobacco smoke (Yauk et al. 2007).
Diethylstilbestrol (DES)
Decades ago, human males exposed in utero to DES were found to have excess occurrence of urogenital malformations, including hypospadias (Henderson et al. 1976; Vessey et al. 1983; Whitehead and Leiter 1981), and elevated prevalence of hypospadias was subsequently reported among unexposed boys whose mothers had been exposed in utero to DES (Brouwers et al. 2006; Klip et al. 2002; Palmer et al. 2005; Pons et al. 2005). More recently, in a study addressing potential competing effects of measured environmental and genotypic factors, a notable excess of hypospadias was reported among unexposed grandsons of women who had been exposed in utero to DES (Kalfa et al. 2011). One postulated mechanism for these observations is that epigenetic changes in the androgen receptor gene are induced in primordial germ cells of DES-exposed female fetuses at the time that the reproductive system is forming, and subsequently transmitted across generations to affected sons and grandsons (Kalfa et al. 2011). To our knowledge, this mechanism has not been examined at the molecular level.
Methodological challenges
Exposure assessment
The long induction periods between exposure and onset of many diseases that epigenetic mechanisms are proposed to mediate present notable challenges to accurate assessment of environmental exposures. Most exposures can change over time, complicating both accurate recall and assessment by short-lived biomarkers. Distinguishing exposures of limited duration or periods of specific relevance (e.g., in utero) from chronic exposures may lend useful specificity for addressing some research questions. Persistent biomarkers (e.g., bone lead levels), when available, are promising if the specific period of exposure is not crucial. Exposure assessed prospectively and tightly linked to proposed periods of vulnerability of the epigenome (e.g., periods of placental invasion or sex specification in utero) would be ideal. However, this rarely occurs in epidemiologic settings with sufficient power to detect most disease associations. However, estimating exposure–epigenetic state associations may be more feasible.
Exposures of limited duration corresponding to disasters (e.g., famine), availability of regulated or withdrawn compounds (e.g., DES), or therapeutic indications of limited duration (e.g., ART) can sometimes be retrospectively assessed with reasonable accuracy. Cohorts based on such exposures, although rare and often small, may be especially valuable in efforts to elucidate transgenerational processes.
Since assessment of covariates may be equally challenging, there is considerable potential for confounding of exposure–epigenetic state or exposure–disease associations by factors associated with exposure, except in rare circumstances when exposure can be randomly assigned to study participants.
Laboratory technologies
Traditional molecular biological methods of analyzing gene loci often involve amplification through cloning, whole-genome amplification (WGA), or polymerase chain reaction (PCR). All of these techniques erase any existing marks in extracted DNA or chromatin. Therefore, methods have been developed to alter the DNA using methods influenced by the epigenetic state prior to amplification. The three main methods employed are affinity enrichment, restriction digestion, and chemical conversion by sodium bisulfite (Laird 2010). Affinity enrichment is appropriate for most epigenetic marks, generally relying on the use of an antibody specific for the mark of interest to enrich for DNA sequences containing the relevant mark by chromatin immunoprecipitation (ChIP). Affinity enrichment can also be used for DNA methylation, but restriction digestion with methylation-resistant and sensitive restriction enzymes has also seen widespread use. Treatment of denatured DNA with sodium bisulfite can convert unmethylated cytosines to uracil, while leaving methylated cytosines intact, thus converting epigenetic information into a genetic difference. As microarray technology became more widespread in the past decade, restriction enzyme and affinity-based methods became popular. With the development of commercial array platforms and the surging use of next-generation sequencing, bisulfite-based methods are now the most widely used methodology, and are well-suited for the types of biospecimens collected in epidemiologic studies.
Biospecimens
The requirement of relatively large amounts of intact chromatin for ChIP-chip or ChIP-seq has limited the study of histone modifications and other protein-associated epigenetic marks to biospecimens with large amounts of material with intact cells. The vast majority of studies on ChIP have used cultured cell lines. More recently, techniques to analyze small quantities of fresh tissue have been explored (Adli and Bernstein 2011). However, none of the existing techniques for analysis of histone modifications are ready for use on the types of samples that have typically been collected in population-based studies, such as blood, buccal swabs, newborn dried blood spots, or formalin-fixed and paraffin-embedded (FFPE) samples. Therefore, most studies have relied almost exclusively on the analysis of DNA methylation, which can be performed on small quantities of extracted DNA. Even heavily cross-linked and degraded DNA obtained from FFPE samples can be interrogated for the distribution of methylation marks.
Investigation of epigenetic mechanisms requires consideration of specific type or types of cells to be studied, since normal epigenetic state varies between cell types (Wu et al. 2011). For many diseases and exposures, the relevant affected cells or tissue may not be readily accessible or even known. The cell types of greatest relevance are unlikely to be represented in extant biorepositories assembled for genetic research, for which most nucleated cells suffice, making minimally invasive collection a primary consideration. Proxy tissue may in special circumstances be useful following deliberate validation (Talens et al. 2010). Where cells of greatest relevance are collected several concerns remain: there may be ethical barriers to obtaining relevant cells from unaffected control individuals or even from cases; cells may not be sufficiently abundant or amenable to separation from surrounding tissue; and if disease processes alter proportions of normal cells spurious associations between epigenetic state and disease may occur on the basis of normal epigenetic profiles alone (Wu et al. 2011).
Statistical methods
Statisticians have developed various methods for disentangling imprinting, parent-of-origin, and maternal–fetal interaction effects in family data (Ainsworth et al. 2011). These generally entail extensions of the transmission-disequilibrium test, which seeks departures in genotype distributions among affected offspring from expected distributions given their parents’ genotypes. The standard test ignores differences between the two parents, so that aA × aa and aa × aA mating types would be considered interchangeable. However, by adding additional terms to the conditional likelihood to distinguish between maternal and paternal alleles, as well as interactions between maternal and offspring genotypes and by incorporating grandmaternal genotypes, it may be possible to separate these phenomena. Rather than the standard likelihood framework for family-based association studies, Weinberg and colleagues (Vermeulen et al. 2009) have developed a flexible log-linear modeling framework based on the joint distribution of mating types and offspring genotypes and extensions to include grandparental genotypes. One particularly important development is the “pent” design (Mitchell and Weinberg 2005) for studying maternal–fetal interactions, entailing genotypes of a mother, her offspring, her spouse, and her parents; in this design, paternal grandparents are not needed, as their mating type can be imputed. However, none of these methods directly incorporate measured epigenetic states.
Development of statistical methods for analysis of DNA methylation, the most frequently measured epigenetic mark, has been dispersed across applications using different technologies. Although a large body of work is starting to accumulate for microarrays, there are still three major platforms in use, each with their own statistical analysis methods (see Siegmund 2011 for a review). Cross-platform adaptation of some methods (e.g., subset quantile normalization Aryee et al. 2011) is an area of current investigation.
Most methods for biological analyses focus on either searching for new disease subtypes through cluster analysis (Clifford et al. 2011; Siegmund et al. 2004), or looking for areas of differential DNA methylation between samples under different conditions (Jaffe et al. 2012a; Langevin et al. 2011). Sometimes differential DNA methylation is defined by differences in variation, rather than a shift in the mean (Jaffe et al. 2012a).
There has been relatively little emphasis on modeling epigenetic state as an intermediate variable. In one such example, however, Siegmund et al. (2006) used a finite mixture model to estimate the association between exposure and latent disease subtype measured by DNA methylation profiles and compared these results with a simpler two-phase approach, first clustering the DNA methylation data and then relating these clusters to exposure using logistic regression.
Several authors (Furrow et al. 2011; Slatkin 2009; Tal et al. 2010) have developed models of epigenetic inheritance that allow epigenetic state to be reset between generations, as well as environmental induction and environment-sensitive modifications. They have shown that variation in either the environmental or the epigenetic state could produce high heritability, comparable to the heritability expected from purely genetic effects. These papers, however, are focused more on theoretical assessment of how much of the “missing heritability” could be due to epigenetics rather than providing methods for analysis of epidemiologic data. We therefore briefly outline a general framework that could be used for this latter purpose. Details and supporting simulation studies will be described completely elsewhere.
One approach could be to treat an individual’s epigenetic state as a time-dependent latent process, μ i (t), which is only partially observed at particular time points t k by flawed assays M ik . Using standard measurement error modeling approaches, one might then treat the latent process as being determined by one’s constitutional genotype G i and exposure history E i (t), through a linear longitudinal random-effects model, with the latent process in turn being a risk factor for disease state Y i (t), through a standard survival analysis model (see, for example, Elashoff et al. 2007) for similar latent process methods applied to longitudinal data on CD4 cell counts in relation to AIDS incidence). To address the high-dimensional nature of epigenetic data, one could use the kinds of cluster-analysis techniques described by Siegmund et al. (2006), treating the cluster rather than individual epigenetic marks, as a latent risk factor for disease (see Molitor et al. 2003 for an example of similar latent cluster methods applied to haplotype associations).
To address transgenerational phenomena, we propose an extension of this approach illustrated in Fig. 2. In this model, grandparental genetic and environmental effects (G mm ,E mm etc.) are transmitted to the parents via transmitted epigenotypes (G mm * etc.), which are modified by the parents’ environments and in turn transmitted to the offspring generation to determine their phenotypes, in combination with their own exposures at the relevant developmental stage.
Extending to genome-wide scale
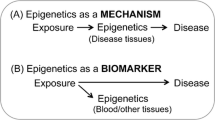
Much of the early work on the role of epigenetics in disease has focused either on global methylation, differentially methylated regions associated with several imprinted loci, or promoter regions of selected candidate genes. Now, facilitated by the advent of high-density methylation arrays like the Illumina Infinium Beadarray that can interrogate up to 480K CpG sites, the kinds of agnostic genome-wide scans that have become popular for investigating germline genetic variation [genome-wide association scans (GWAS)] are likely to become widely used for epigenetics as well. There are two forms such studies could take. The first is a genome-wide scan for epigenetic variation in relation to disease risk—what has recently been called an epigenome-wide association scan (EWAS) (Rakyan et al. 2011) (this should not to be confused with the use of the same acronym for exposure-wide association scans, Patel et al. 2010). For example, in an agnostic scan of 92 head and neck cancers and 92 controls, 6 CpG loci were identified (Langevin et al. 2012). Here, the focus was on finding biomarkers that might be useful for early detection rather than for etiologic inference. Case–control designs are commonly used in GWAS studies, wherein measured biomarkers of interest are genotypes that do not change over time and are presumed to be identical in all nucleated cells of each individual. EWAS for etiology are far more challenging for two fundamental reasons. First, use of retrospective case–control designs introduces the possibility of “reverse causation,” wherein the disease or its treatment causes epigenetic changes rather than the reverse. Second, tissue and cell type specificity of epigenetic marks measured in EWAS pose the issues of sample collection and interpretation described in the previous section.
A different example, provided by an EWAS of the effect of smoking on methylation (Breitling et al. 2011), which found lower methylation at a CpG site in F2RL3 in smokers than nonsmokers, avoids the problem of reverse causation, but not of tissue specificity.
The second approach to agnostic genome-wide epigenetic research uses methylation quantitative trait loci (mQTLs). These seek genome-wide SNP variation associated with genome-wide epigenetic state (Bell et al. 2011a) in a manner similar to expression QTL (eQTL) studies. Studies of mQTLs offer the potential to detect both cis-associations (SNPs in or near a gene that are associated with epigenetic state of the same gene) and trans-associations (associations with epigenetic state of genes located at some distance from the SNP or on separate chromosomes). Although the vast majority of associations in HapMap cell lines studied by Bell et al. were of the cis type, they found a trans association of a SNP in the DIP2B gene (thought to be involved in DNA methylation) with the first principal component of genome-wide methylation.
EWASs pose novel statistical challenges for peak detection that differ from other applications of this techniques (such as for ChIPseq data), as summarized by Jaffe et al. (2012b). Not only a larger number of individuals studied, but the correlation structure from adjacent probes on the microarray is more complex, peaks are expected to be more variable in size and shape and in certain applications more numerous (requiring different approaches for multiple comparisons) and there can be more serious measurement error problems. Jaffe et al. propose a novel “bump hunting” technique to address such problems. Another challenge concerns the problem of reverse causation mentioned above. In studies of the association of biomarkers with disease, the technique of Mendelian randomization (Davey Smith and Ebrahim 2004) has received much attention as a way of overcoming reverse causation and uncontrolled confounding by using a gene as an instrumental variable (Greenland 2000) to assess the causal effect of the biomarker on disease risk. Relton and Davey Smith (2012) have proposed a novel two-step extension of this idea for methylation studies, using two genes as instrumental variables, one to estimate the exposure–methylation association, the other to estimate the methylation–disease association.
Epidemiological opportunities
Despite the wealth of research on the role of epigenetics in disease, there has been much less activity on the environmental determinants of epigenetic changes and their role of epigenetic state in mediating exposure–response relationships, either within individuals or across generations. Rakyan et al. (2011) provide an excellent review of some of the design issues in epigenetic epidemiology. Here we focus on some opportunities for further epidemiologic study in this area.
Case–control and cohort studies
Studies of the genetic determinants of disease, whether based on candidate site hypotheses, pathway-based hypotheses, or GWAS technology, have utilized cross-sectional studies in what is conceptually a case–control approach, since the “exposures” in question were genotypes presumed to be immutable. Analogous studies of disease outcomes in light of epigenetic alterations, however, are fundamentally flawed, not only because earlier causal events could always be responsible for observed epigenetic change, but also because any stage in the pathophysiology of the disease and/or any treatment prior to sample collection could itself generate epigenetic change. De novo cohort studies of the genetic determinants of disease require biosamples at the outset, are slow to yield results, and often prohibitively expensive, because the rare outcomes of greatest interest can only be assessed if the genomic subsets are very large. Cohort studies of the causation of endpoint phenotypes or even intermediary risk factors by epigenetic characteristics are likely to be just as difficult. Moreover, observational studies of human epigenetic events as usually conducted are confounded by age, genomic characteristics, and the earlier environmental milieu, including the prepartum component.
One can, however, study epigenetic alterations as outcomes, and by so doing, come to understand their role as intermediary agents between disease outcomes and the toxic agents known to cause them. Cross-sectional studies of cells from exposed and unexposed individuals are interpretable under the assumption that the epigenetic alterations are unlikely to have produced the behavior that led to the toxic exposure. Over the long term, comparison between the epigenetic experiences of exposed and unexposed cohorts will require repeated epigenetic characterization of persons with particular toxic exposures or particular characteristics, such as jobs that stand as surrogate measures of such exposure.
Twin studies
In contrast to observational studies of singletons, twin studies can provide several advantages for studies of epigenetics (Bell and Saffery 2012). Monozygotic twins are in effect matched on age, complete genome, and childhood environment and cultural milieu. By contrast, even monozygotic twins can be considered only partly matched on intrauterine environment. Because epigenetic events occur at various points throughout gestation, epigenetic state at birth is probably influenced by chorionicity and possibly as well by characteristics of the amnion and the anatomy of implantation.
Differences between the global and gene-specific epigenetic states of adult identical twins have been described in numerous studies (Bell et al. 2012; Bocklandt et al. 2011; Boks et al. 2009; Fraga et al. 2005; Heijmans et al. 2007), suggesting that epigenetic state changes with age, or more accurately, with cohort, since these were all different people. Methylation state at some CpG sites is highly predictive, as shown in a recent study that identified numerous methyl marks in DNA isolated from saliva to be highly associated with age. In a validation set of study participants, methylation state at only two of the identified CpG sites explained 73 % of the variance in age over a 5-decade range (Bocklandt et al. 2011). Such epigenetic marks may prove useful not only in understanding biological process associated with aging, but also as tools for classifying participants in research into biologically based age groups. For example, individuals with large differences between epigenetically predicted age and chronological age might be considered to have unusual physiologic state or bio-age.
However, unless age-specific epigenetic states are demonstrated in the same twins studied across different ages, as has been done in singletons (Bjornsson et al. 2008), apparent effects of age on epigenetic state may be confounded by birth cohort-specific differences in exposure history. In a study of identical twin children observed at both ages 5 and 10, prevalence of DNA methylation at selected promoter sites changed measurably over time (Wong et al. 2010), as did prevalence of more extreme skewing of X chromosome inactivation within some female pairs (Wong et al. 2011). Observed concordance between co-twins of changes in such measures provide information regarding factors that may influence epigenetic changes that is not available from singletons. For example, observing little concordance of within-pair changes in X chromosome inactivation, Wong et al. (2011) concluded that alterations in these patterns between 5 and 10 years of age are unlikely to be influenced by factors shared by twins. Establishment of cohorts to be periodically monitored for epigenetic changes should provide further documentation of age-specific changes (Saffery et al. 2011).
Identical twins discordant for both epigenetic state and a putatively toxic exposure or adverse circumstance can therefore be used to tentatively identify factors that cause or facilitate epigenetic change. For example, in a study comparing members of monozygotic dichorionic twin pairs, DNA from placental tissue of smaller co-twins was found to have lower prevalence of promoter DNA methylation at the human endogenous retroviral family W Env(C7) member 1 gene (HERVWE1). Moreover, mRNA and protein encoded by HERVWE1 was more highly expressed in placental tissue of smaller co-twins (Gao et al. 2012).
Studies identifying differences in epigenetic state between identical twin pairs discordant for a trait or disease phenotype can raise provocative etiologic questions. However, as in case–control studies of phenotypic outcomes in general, these studies must be interpreted with caution, particularly if the chronology of relevant exposures, occurrence of epigenetic changes, and onset of pathophysiologic processes is unclear, if the variable locus is not known to function in the pathogenic pathway, or if the disease pathology is not manifested in the tissue in which epigenetic state is measured. Such provocative questions have been raised by studies of trait-discordant monozygotic twins in relation to differential X-inactivation in the context of bipolar disorder (Rosa et al. 2008) and primary biliary cirrhosis (Mitchell et al. 2011), and differential DNA methylation at specific loci in the context of a caudal duplication anomaly (Oates et al. 2006), bipolar disorder (Dempster et al. 2011; Kuratomi et al. 2008), schizophrenia (Dempster et al. 2011), and BRCA1-related malignancy (Galetzka et al. 2012). Global changes in methylation, to differing degrees, have been reported in identical twins discordant for psychometric evidence of aberrant personality (Kaminsky et al. 2008), Alzheimer’s disease (Mastroeni et al. 2009), systemic lupus erythematosus (Javierre et al. 2010), autism (Nguyen et al. 2010a), and inflammatory bowel disease (Harris et al. 2012). A comprehensive global search for differences between (only three pairs of) twins discordant for multiple sclerosis was unsuccessful, possibly because peripheral white blood cells were used (Baranzini et al. 2010). Differences in gene expression between trait-discordant identical twins, presumably reflecting epigenetic differences, have been observed for rheumatoid arthritis (Haas et al. 2006), bipolar disorder (Matigian et al. 2007), schizophrenia (Kakiuchi et al. 2008), type 1 diabetes (Beyan et al. 2010), diverse systemic autoimmune conditions (O’Hanlon et al. 2011), and psoriasis (Gervin et al. 2012).
Twins have also been used to suggest the existence of heritable determinants of epigenetic change. The traditional design is based on the comparison of proband-wise concordance between monozygotic twins with that between like-sex dizygotic twins, separating the variance attributable to the environment from that attributable to the additional half of the genome held in common between monozygotic twins. Reservations about conclusions from such studies are based on two considerations. The first concerns the assumption that the sharing of the monozygotic twins’ environment is identical to that of dizygotic twins—an assumption necessary for heritability to be estimable from twin studies. This assumption is suspect in adulthood, since dizygotic twins tend to diverge geographically and socially to a greater extent than do monozygotic twins, and since any monozygotic twin is more likely than a dizygotic twin to perceive and adopt a behavior seen to be advantageous to the co-twin. This is compounded by the second concern, namely the inevitable variability in the twin concordance of a phenotype that is also determined by a geographically, chronologically, or socially variable environmental exposure. Thus, the reliability of any such estimate is highest for phenotypes that are not confounded by age, time, and place, and especially for those of young children that have no behavioral determinants.
Moreover, when comparisons of twin concordance are used to estimate the heritability of an epigenetic trait, necessary caveats are more numerous. Heritability of epigenetic traits is clearly tissue specific, and likely to be locus specific as well. Because of the influence of the intrauterine environment and the fact that they exchange blood cells, monochorionic identical twins cannot be considered comparable to dizygotic twins, and comparisons should be based only on dichorionic monozygotic pairs. Irrespective of chorionicity, identical twins begin life as a single zygote, restricting the number of early cell-days at risk of independent epigenetic change relative to the separate zygotes of dizygotic twins.
Initial attempts at demonstrating heritability at specific loci by these methods have had some limited success (Boks et al. 2009; Breton et al. 2011c; Coolen et al. 2011; Heijmans et al. 2007; Schneider et al. 2010), but investigations of larger portions of the epigenome in specific tissues have shown only a marginal increase in epigenetic similarity between the members of monozygotic twin pairs, compared with dizygotic twin pairs (Gervin et al. 2011; Ollikainen et al. 2010).
Multigenerational studies
In the remainder of this section, we discuss a few of the unique exposure settings that might be useful for exploring the role of epigenetics in apparent transgenerational exposure phenomena.
Famine cohorts
Testis cancer incidence has been increasing for decades, and age–period–cohort analyses consistently demonstrate that birth year is an important determinant of testicular cancer risk. Based on these findings, exposure to unidentified environmental causes is postulated to be increasing, with observed birth cohort effects attributed to early action of such exposures. The singular exception to this pattern is a dip in incidence among men born in Europe during World War II (Moller et al. 1995). Postulated epigenetic mechanisms related to energy intake in the in utero and perinatal periods warrant investigation among individuals gestated during famine and their descendents.
Child health and development studies
The California Child Health and Development Studies (Cohn 2011; van den Berg et al. 1988) were launched between 1959 and 1967 with the enrollment of a cohort of about 20,000 pregnancies, with follow-up of the parents and offspring through the present day. Biological specimens were provided by both parents and the child at the time of enrollment, along with limited questionnaire information about grandparental exposures, and longitudinal samples have also been obtained. As these children are now in their 50s, many have themselves had children, who have also been enrolled in the study, and grandchildren are already starting to be born. This cohort thus represents a unique opportunity to study transgenerational epigenetic phenomena, such as their studies of maternal exposures to DDT.
Diethylstilbestrol offspring
DES is regarded as a potential environmental endocrine disruptor by many researchers because this compound binds steroid hormone receptors and has only exogenous sources. Because exposure occurred only in relatively controlled clinic settings, cohorts of exposed individuals could in theory provide an opportunity to trace any epigenetic effects of exposure among themselves and their descendents to the restricted time periods of their pregnancies or gestation before administration to pregnant women ended in the 1970s. Exposure and comparison group data of varying quality are or could be made available from several sources, the highest quality in theory being participants in trials of DES efficacy conducted in the 1950s (Dieckmann et al. 1953; Ferguson 1953), although sample sizes were limited. Nonetheless, the numerous case–control sets, DES-exposed cohorts, and interest groups that subsequently enrolled large numbers of exposed men and women to monitor and investigate health effects of exposed individuals (Swan 2000) could prove to be valuable resources for investigation of potential epigenetic and transgenerational effects of DES among exposed individuals and their descendents. Initial proof-of-concept studies could include investigation of epigenetic marks postulated to have been disrupted by DES exposures.
Assisted reproductive technologies
In industrialized countries 1–4 % of neonates are now conceived by ART. Intensive surveillance of long-term health of these children has revealed somewhat increased occurrence of numerous disorders (Savage et al. 2011). However, a desirable next step in this research—determining whether ART procedures or indications for these procedures (parents’ infertility or subfertility) account for excess risk—is severely limited by paucity of data on children with a history of only one of these factors. Epigenetic mechanisms are commonly postulated for both ART and parental origins of several ART-associated conditions, including model imprinting disorders and IUGR. Therefore, new studies aiming to trace candidate epigenetic marks from relevant cells of the parent (e.g., father’s sperm) to those of affected ART-conceived children (e.g., placenta of IUGR infants or somatic cells of children with Prader-Willi syndrome) seems a logical first step to separating treatment effects from indication. More generally, studies comparing the epigenetic state of relevant cells from children conceived by ART with that of like-cell types from naturally conceived children may provide additional insights regarding conditions in early development, particularly conditions over-represented among children conceived by ART. Ongoing surveillance will doubtlessly reveal whether later onset conditions are added to this list as cohorts of children conceived by ART achieve ages at risk of diseases of adulthood. A logical extension of this surveillance would be to determine among descendents the presence of relevant phenotypes and epigenetic marks.
Conclusions
Noteworthy experimental results, taken together, indicate that epigenetic processes could plausibly mediate effects of environmental exposures to influence disease susceptibility in mammals, since (1) specific exposures induced measurable changes in epigenetic state, (2) new epigenetic state was shown to persist after cessation of exposure, and (3) measureable phenotype was associated with exposure-related changes in epigenetic state. A fourth element, exposure-induced alteration of epigenetic marks of germ cell lineage DNA, seems necessary for transgenerational transmission of environmentally induced epigenetic changes. On this background, we interpret observational human data illustrating associations of exposures to epigenetic state and epigenetic state to disease to indicate that epigenetic mechanisms may plausibly mediate exposure effects on a wide range of human diseases.
Perhaps the greatest significance of this line of research is the potential for designing novel interventions articulated by Dolinoy and Jirtle (2008):
unlike genetic mutations, epigenetic profiles are potentially reversible. Therefore, epigenetic approaches for prevention and treatment, such as nutritional supplementation and/or pharmaceutical therapies may be developed to counteract negative epigenomic profiles.
As an example, Huang et al. (2012) have developed a therapy for reactivating the Ube3a allele that is epigenetically silenced in Angelman syndrome. However, before we begin to realize this translational potential, much research along parallel tracks needs to be conducted.
We need to better understand basic epigenetic mechanisms that operate during selected periods when the epigenome may be particularly vulnerable to environmental exposures (e.g., prenatal and early postnatal periods, childhood, puberty) as well as mechanisms that routinely maintain proper epigenetic states. We also need to understand mechanisms relevant to cumulative effects of exposure and associations with aging, since duration of exposure and aging are co-linear. A better understanding of basic mechanisms at different time points in life would guide genetic epidemiology studies in selecting environmental exposures that are more likely to affect epigenetic processes, identify the most relevant periods of exposure, identify biomarkers of exposure, and suggest biological intermediates to study.
It may be helpful to more deliberately deconstruct the steps whereby an environmental exposure may affect risk of disease via epigenetic mechanisms. For example, studies of the association of environmental exposures with selected epigenetic events as endpoints (rather than intermediates in a complex pathway) may be fruitful and will likely help us better conceive of studies to elucidate an entire pathway, from an exposure acting at least in part through epigenetic mechanisms to disease risk.
As we proceed, it will be important to consider seriously the environmental exposures to study, since we will want to select the most promising candidate exposures (those most likely to affect epigenetic processes), measuring these exposures at relevant time periods and with reasonable accuracy. Also, we cannot ignore genetic influences on epigenetic processes. We see above that studies of the epigenetic state of specific candidate genes are yielding interesting results and more such studies are warranted. Given limited knowledge of this genetic–epigenetic interface, to complement the candidate gene approach, integrative genomics approaches should be contemplated in order to combine GWAS data with epigenomic data. Clearly, statistical and bioinformatics approaches will be required to enable the efficient conduct of these analyses, especially as we expand to genome-wide scale. Similarly, research on feasible, efficient study designs, akin to current research on optimum designs for sequencing-based studies, will be needed. Finally, as with other emerging fields (e.g., the standardization of testing criteria for microsatellite instability or MSI), standardization of terminology and testing protocols will facilitate future communication of hypotheses, scientific approaches, and results.
Abbreviations
- ART:
-
Assisted reproductive technologies
- ASM:
-
Allele-specific DNA methylation
- ChIP:
-
Chromatin immunoprecipitation
- CIMP:
-
CpG island methylator phenotype
- CpG:
-
Cytosine-phosphate-guanine dinucleotide
- CRC:
-
Colorectal cancer
- DES:
-
Diethylstilbestrol
- feNO:
-
Fractional exhaled nitric oxide
- FFPE:
-
Formalin-fixed paraffin-embedded
- HDAC:
-
Histone deacetylases
- iNOS:
-
Inducible nitric oxide synthase
- IUGR:
-
Intra-uterine growth restriction
- IVF:
-
In vitro fertilization
- PBMCs:
-
Peripheral blood mononuclear cells
- PTS:
-
Maternal smoking during pregnancy
- SNP:
-
Single nucleotide polymorphism
References
Adli M, Bernstein BE (2011) Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc 6:1656–1668
Ainsworth HF, Unwin J, Jamison DL, Cordell HJ (2011) Investigation of maternal effects, maternal-fetal interactions and parent-of-origin effects (imprinting), using mothers and their offspring. Genet Epidemiol 35:19–45
Akbarian S (2010) Epigenetics of schizophrenia. Curr Top Behav Neurosci 4:611–628
Andersen AN, Pinborg A, Loft A (2008) Neonatal outcome in singletons conceived after ART. Lancet 372:694–695
Anway MD, Skinner MK (2008) Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate 68:517–529
Anway MD, Cupp AS, Uzumcu M, Skinner MK (2005) Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308:1466–1469
Apostolidou S, Abu-Amero S, O’Donoghue K, Frost J, Olafsdottir O, Chavele KM, Whittaker JC, Loughna P, Stanier P, Moore GE (2007) Elevated placental expression of the imprinted PHLDA2 gene is associated with low birth weight. J Mol Med (Berlin) 85:379–387
Arita A, Niu J, Qu Q, Zhao N, Ruan Y, Nadas A, Chervona Y, Wu F, Sun H, Hayes RB, Costa M (2011) Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environ Health Perspect 120:198–203
Aryee MJ, Wu Z, Ladd-Acosta C, Herb B, Feinberg AP, Yegnasubramanian S, Irizarry RA (2011) Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics 12:197–210
Baccarelli A, Bollati V (2009) Epigenetics and environmental chemicals. Curr Opin Pediatr 21:243–251
Baccarelli A, Wright RO, Bollati V, Tarantini L, Litonjua AA, Suh HH, Zanobetti A, Sparrow D, Vokonas PS, Schwartz J (2009) Rapid DNA methylation changes after exposure to traffic particles. Am J Respir Crit Care Med 179:572–578
Badcock C, Crespi B (2008) Battle of the sexes may set the brain. Nature 454:1054–1055
Baranzini SE, Mudge J, van Velkinburgh JC, Khankhanian P, Khrebtukova I, Miller NA, Zhang L, Farmer AD, Bell CJ, Kim RW, May GD, Woodward JE, Caillier SJ, McElroy JP, Gomez R, Pando MJ, Clendenen LE, Ganusova EE, Schilkey FD, Ramaraj T, Khan OA, Huntley JJ, Luo S, Kwok PY, Wu TD, Schroth GP, Oksenberg JR, Hauser SL, Kingsmore SF (2010) Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464:1351–1356
Bariol C, Suter C, Cheong K, Ku SL, Meagher A, Hawkins N, Ward R (2003) The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol 162:1361–1371
Barker DJ, Gluckman PD, Godfrey KM, Harding JE, Owens JA, Robinson JS (1993) Fetal nutrition and cardiovascular disease in adult life. Lancet 341:938–941
Bayarsaihan D (2011) Epigenetic mechanisms in inflammation. J Dent Res 90:9–17
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11:726–734
Begemann M, Spengler S, Kanber D, Haake A, Baudis M, Leisten I, Binder G, Markus S, Rupprecht T, Segerer H, Fricke-Otto S, Muhlenberg R, Siebert R, Buiting K, Eggermann T (2011) Silver-Russell patients showing a broad range of ICR1 and ICR2 hypomethylation in different tissues. Clin Genet 80:83–88
Bell JT, Saffery R (2012) The value of twins in epigenetic epidemiology. Int J Epidemiol 41:140–150
Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK (2011a) DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12:R10
Bell O, Tiwari VK, Thoma NH, Schubeler D (2011b) Determinants and dynamics of genome accessibility. Nat Rev Genet 12:554–564
Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, Shin SY, Dempster EL, Murray RM, Grundberg E, Hedman AK, Nica A, Small KS, Dermitzakis ET, McCarthy MI, Mill J, Spector TD, Deloukas P (2012) Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet 8:e1002629
Benchaib M, Ajina M, Lornage J, Niveleau A, Durand P, Guerin JF (2003) Quantitation by image analysis of global DNA methylation in human spermatozoa and its prognostic value in in vitro fertilization: a preliminary study. Fertil Steril 80:947–953
Benchaib M, Braun V, Ressnikof D, Lornage J, Durand P, Niveleau A, Guerin JF (2005) Influence of global sperm DNA methylation on IVF results. Hum Reprod 20:768–773
Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CPE, van Dijk CM, Tollenaar RAEM, Van Den Berg D, Laird PW (2011) Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina–associated domains. Nat Genet 43 (in press)
Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, Van Den Berg D, Laird PW (2012) Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 44:40–46
Beyan H, Drexhage RC, van der Heul Nieuwenhuijsen L, de Wit H, Padmos RC, Schloot NC, Drexhage HA, Leslie RD (2010) Monocyte gene-expression profiles associated with childhood-onset type 1 diabetes and disease risk: a study of identical twins. Diabetes 59:1751–1755
Birney E, Lieb JD, Furey TS, Crawford GE, Iyer VR (2010) Allele-specific and heritable chromatin signatures in humans. Hum Mol Genet 19:R204–R209
Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP (2008) Intra-individual change over time in DNA methylation with familial clustering. JAMA 299:2877–2883
Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, Vilain E (2011) Epigenetic predictor of age. PLoS One 6:e14821
Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet 30:73–76
Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA (2009) The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One 4:e6767
Bollati V, Baccarelli A (2010) Environmental epigenetics. Heredity 105:105–112
Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, Byun HM, Jiang J, Marinelli B, Pesatori AC, Bertazzi PA, Yang AS (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res 67:876–880
Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V, Tarantini L, Cantone L, Schwartz J, Bertazzi PA, Baccarelli A (2010) Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 118:763–768
Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H (2011) Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet 88:450–457
Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H (2012) Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J [Epub ahead of print]
Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD (2009) Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med 180:462–467
Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD (2011a) DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med 184:191–197
Breton CV, Salam MT, Gilliland FD (2011b) Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics 6:895–898
Breton CV, Salam MT, Gilliland FD (2011c) Heritability and role for the environment in DNA methylation in AXL receptor tyrosine kinase. Epigenetics 6:895–898
Brouwers MM, Feitz WF, Roelofs LA, Kiemeney LA, de Gier RP, Roeleveld N (2006) Hypospadias: a transgenerational effect of diethylstilbestrol? Hum Reprod 21:666–669
Bruce KD, Hanson MA (2010) The developmental origins, mechanisms, and implications of metabolic syndrome. J Nutr 140:648–652
Burdge GC, Slater-Jefferies J, Torrens C, Phillips ES, Hanson MA, Lillycrop KA (2007) Dietary protein restriction of pregnant rats in the F0 generation induces altered methylation of hepatic gene promoters in the adult male offspring in the F1 and F2 generations. Br J Nutr 97:435–439
Byun HM, Siegmund KD, Pan F, Weisenberger DJ, Kanel G, Laird PW, Yang AS (2009) Epigenetic profiling of somatic tissues from human autopsy specimens identifies tissue- and individual-specific DNA methylation patterns. Hum Mol Genet 18:4808–4817
Campion J, Milagro FI, Martinez JA (2009) Individuality and epigenetics in obesity. Obes Rev 10:383–392
Cantone L, Nordio F, Hou L, Apostoli P, Bonzini M, Tarantini L, Angelici L, Bollati V, Zanobetti A, Schwartz J, Bertazzi PA, Baccarelli A (2011) Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ Health Perspect 119:964–969
Carrell DT, Hammoud SS (2010) The human sperm epigenome and its potential role in embryonic development. Mol Hum Reprod 16:37–47
Chan TL, Yuen ST, Kong CK, Chan YW, Chan AS, Ng WF, Tsui WY, Lo MW, Tam WY, Li VS, Leung SY (2006) Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 38:1178–1183
Chanda S, Dasgupta UB, Guhamazumder D, Gupta M, Chaudhuri U, Lahiri S, Das S, Ghosh N, Chatterjee D (2006) DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol Sci 89:431–437
Christensen BC, Marsit CJ (2011) Epigenomics in environmental health. Front Genet 2:84
Ciappio ED, Mason JB, Crott JW (2011) Maternal one-carbon nutrient intake and cancer risk in offspring. Nutr Rev 69:561–571
Clifford H, Wessely F, Pendurthi S, Emes RD (2011) Comparison of clustering methods for investigation of genome-wide methylation array data. Front Genet 2:88
Cohn BA (2011) Developmental and environmental origins of breast cancer: DDT as a case study. Reprod Toxicol 31:302–311
Coolen MW, Statham AL, Qu W, Campbell MJ, Henders AK, Montgomery GW, Martin NG, Clark SJ (2011) Impact of the genome on the epigenome is manifested in DNA methylation patterns of imprinted regions in monozygotic and dizygotic twins. PLoS One 6:e25590
Cortessis VC (2009) Imprinting errors and in vitro fertilization. In: Carrell DT, Racowski C, Van Voorhis B, Schlegel P (eds) Biennial review of infertility, vol 1. Springer/Humana, Europe, pp 239–246
Cortessis VK, Siegmund K, Houshdaran S, Laird PW, Sokol RZ (2011) Repeated assessment by high-throughput assay demonstrates that sperm DNA methylation levels are highly reproducible. Fertil Steril 96:1325–1330
Cropley JE, Suter CM, Beckman KB, Martin DI (2006) Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Nat Acad Sci 103:17308–17312
Curtin K, Slattery ML, Samowitz WS (2011) CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int 2011:902674
D’Angelo DV, Whitehead N, Helms K, Barfield W, Ahluwalia IB (2011) Birth outcomes of intended pregnancies among women who used assisted reproductive technology, ovulation stimulation, or no treatment. Fertil Steril 96(314–320):e2
Davey Smith G, Ebrahim S (2004) Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 33:30–42
Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Kalidindi S, Picchioni M, Kravariti E, Toulopoulou T, Murray RM, Mill J (2011) Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet 20:4786–4796
Dieckmann WJ, Davis ME, Rynkiewicz LM, Pottinger RE (1953) Does the administration of diethylstilbestrol during pregnancy have therapeutic value? Am J Obstet Gynecol 66:1062–1081
Diergaarde B, Braam H, van Muijen GN, Ligtenberg MJ, Kok FJ, Kampman E (2003) Dietary factors and microsatellite instability in sporadic colon carcinomas. Cancer Epidemiol Biomarkers Prev 12:1130–1136
Diplas AI, Lambertini L, Lee MJ, Sperling R, Lee YL, Wetmur J, Chen J (2009) Differential expression of imprinted genes in normal and IUGR human placentas. Epigenetics 4:235–240
Dolinoy DC, Jirtle RL (2008) Environmental epigenomics in human health and disease. Environ Mol Mutagen 49:4–8
Dolinoy DC, Huang D, Jirtle RL (2007a) Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Nat Acad Sci 104:13056–13061
Dolinoy DC, Weidman JR, Jirtle RL (2007b) Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol 23:297–307
Durham AL, Wiegman C, Adcock IM (2011) Epigenetics of asthma. Biochim Biophys Acta 1810:1103–1109
Eaton AM, Sandler R, Carethers JM, Millikan RC, Galanko J, Keku TO (2005) 5,10-methylenetetrahydrofolate reductase 677 and 1298 polymorphisms, folate intake, and microsatellite instability in colon cancer. Cancer Epidemiol Biomarkers Prev 14:2023–2029
El Hajj N, Zechner U, Schneider E, Tresch A, Gromoll J, Hahn T, Schorsch M, Haaf T (2011) Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex Dev 5:60–69
Elashoff RM, Li G, Li N (2007) An approach to joint analysis of longitudinal measurements and competing risks failure time data. Stat Med 26:2813–2835
El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, Cooper ME, Brownlee M (2008) Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205:2409–2417
Enokida H, Shiina H, Urakami S, Terashima M, Ogishima T, Li LC, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Igawa M, Dahiya R (2006) Smoking influences aberrant CpG hypermethylation of multiple genes in human prostate carcinoma. Cancer 106:79–86
Estecio MR, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, Oki Y, Kondo Y, Jelinek J, Shen L, Hartung H, Aplan PD, Czerniak BA, Liang S, Issa JP (2010) Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res 20:1369–1382
Faulk C, Dolinoy DC (2011) Timing is everything: the when and how of environmentally induced changes in the epigenome of animals. Epigenetics 6:791–797
Feinberg AP (2007) Phenotypic plasticity and the epigenetics of human disease. Nature 447:433–440
Feltus FA, Lee EK, Costello JF, Plass C, Vertino PM (2006) DNA motifs associated with aberrant CpG island methylation. Genomics 87:572–579
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science 330:622–627
Ferguson JH (1953) Effect of stilbestrol on pregnancy compared to the effect of a placebo. Am J Obstet Gynecol 65:592–601
Ferguson-Smith AC (2011) Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 12:565–575
Fields PE, Kim ST, Flavell RA (2002) Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol 169:647–650
Filipponi D, Feil R (2009) Perturbation of genomic imprinting in oligozoospermia. Epigenetics 4:27–30
Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, Gonzalez K, Santella RM, Terry MB (2011) Prenatal smoke exposure and genomic DNA methylation in a multiethnic birth cohort. Cancer Epidemiol Biomarkers Prev 20:2518–2523
Fortier AL, Lopes FL, Darricarrere N, Martel J, Trasler JM (2008) Superovulation alters the expression of imprinted genes in the midgestation mouse placenta. Hum Mol Genet 17:1653–1665
Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Nat Acad Sci 102:10604–10609
Furrow RE, Christiansen FB, Feldman MW (2011) Environment-sensitive epigenetics and the heritability of complex diseases. Genetics 189:1377–1387
Galetzka D, Hansmann T, El Hajj N, Weis E, Irmscher B, Ludwig M, Schneider-Raetzke B, Kohlschmidt N, Beyer V, Bartsch O, Zechner U, Spix C, Haaf T (2012) Monozygotic twins discordant for constitutive BRCA1 promoter methylation, childhood cancer and secondary cancer. Epigenetics 7:47–54
Gao Y, He Z, Wang Z, Luo Y, Sun H, Zhou Y, Huang L, Li M, Fang Q, Jiang S (2012) Increased expression and altered methylation of HERVWE1 in the human placentas of smaller fetuses from monozygotic, dichorionic, discordant twins. PLoS One 7:e33503
Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, Kucera KS, Willard HF, Myers RM (2011) Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS Genet 7:e1002228
Gervin K, Hammero M, Akselsen HE, Moe R, Nygard H, Brandt I, Gjessing HK, Harris JR, Undlien DE, Lyle R (2011) Extensive variation and low heritability of DNA methylation identified in a twin study. Genome Res 21:1813–1821
Gervin K, Vigeland MD, Mattingsdal M, Hammero M, Nygard H, Olsen AO, Brandt I, Harris JR, Undlien DE, Lyle R (2012) DNA methylation and gene expression changes in monozygotic twins discordant for psoriasis: identification of epigenetically dysregulated genes. PLoS Genet 8:e1002454
Gordon L, Joo JH, Andronikos R, Ollikainen M, Wallace EM, Umstad MP, Permezel M, Oshlack A, Morley R, Carlin JB, Saffery R, Smyth GK, Craig JM (2011) Expression discordance of monozygotic twins at birth: effect of intrauterine environment and a possible mechanism for fetal programming. Epigenetics 6:579–592
Greenland S (2000) An introduction to instrumental variables for epidemiologists. Int J Epidemiol 29:722–729
Guo L, Choufani S, Ferreira J, Smith A, Chitayat D, Shuman C, Uxa R, Keating S, Kingdom J, Weksberg R (2008) Altered gene expression and methylation of the human chromosome 11 imprinted region in small for gestational age (SGA) placentae. Dev Biol 320:79–91
Haas CS, Creighton CJ, Pi X, Maine I, Koch AE, Haines GK, Ling S, Chinnaiyan AM, Holoshitz J (2006) Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthr Rheum 54:2047–2060
Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, Olshen A, Ballinger T, Zhou X, Forsberg KJ, Gu J, Echipare L, O’Geen H, Lister R, Pelizzola M, Xi Y, Epstein CB, Bernstein BE, Hawkins RD, Ren B, Chung WY, Gu H, Bock C, Gnirke A, Zhang MQ, Haussler D, Ecker JR, Li W, Farnham PJ, Waterland RA, Meissner A, Marra MA, Hirst M, Milosavljevic A, Costello JF (2010) Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol 28:1097–1105
Harris RA, Nagy-Szakal D, Pedersen N, Opekun A, Bronsky J, Munkholm P, Jespersgaard C, Andersen P, Melegh B, Ferry G, Jess T, Kellermayer R (2012) Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis [Epub ahead of print]
Heijmans BT, Kremer D, Tobi EW, Boomsma DI, Slagboom PE (2007) Heritable rather than age-related environmental and stochastic factors dominate variation in DNA methylation of the human IGF2/H19 locus. Hum Mol Genet 16:547–554
Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH (2008) Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Nat Acad Sci 105:17046–17049
Henderson BE, Benton B, Cosgrove M, Baptista J, Aldrich J, Townsend D, Hart W, Mack TM (1976) Urogenital tract abnormalities in sons of women treated with diethylstilbestrol. Pediatrics 58:505–507
Herceg Z, Paliwal A (2011) Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res 727:55–61
Hitchins MP, Rapkins RW, Kwok CT, Srivastava S, Wong JJ, Khachigian LM, Polly P, Goldblatt J, Ward RL (2011) Dominantly inherited constitutional epigenetic silencing of MLH1 in a cancer-affected family is linked to a single nucleotide variant within the 5′UTR. Cancer Cell 20:200–213
Ho SM (2010) Environmental epigenetics of asthma: an update. J Allergy Clin Immunol 126:453–465
Houshdaran S, Cortessis VK, Siegmund K, Yang A, Laird PW, Sokol RZ (2007) Widespread epigenetic abnormalities suggest a broad DNA methylation erasure defect in abnormal human sperm. PLoS One 2:e1289
Huang Y, Chang X, Lee J, Cho YG, Zhong X, Park IS, Liu JW, Califano JA, Ratovitski EA, Sidransky D, Kim MS (2011) Cigarette smoke induces promoter methylation of single-stranded DNA-binding protein 2 in human esophageal squamous cell carcinoma. Int J Cancer 128:2261–2273
Huang HS, Allen JA, Mabb AM, King IF, Miriyala J, Taylor-Blake B, Sciaky N, Dutton JW Jr, Lee HM, Chen X, Jin J, Bridges AS, Zylka MJ, Roth BL, Philpot BD (2012) Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature 481:185–189
Hughes LA, van den Brandt PA, de Bruine AP, Wouters KA, Hulsmans S, Spiertz A, Goldbohm RA, de Goeij AF, Herman JG, Weijenberg MP, van Engeland M (2009) Early life exposure to famine and colorectal cancer risk: a role for epigenetic mechanisms. PLoS One 4:e7951
Iacopetta B, Grieu F, Phillips M, Ruszkiewicz A, Moore J, Minamoto T, Kawakami K (2007) Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci 98:1454–1460
Ideraabdullah FY, Abramowitz LK, Thorvaldsen JL, Krapp C, Wen SC, Engel N, Bartolomei MS (2011) Novel cis-regulatory function in ICR-mediated imprinted repression of H19. Developmental biology 355:349–357
Jaffe AE, Feinberg AP, Irizarry RA, Leek JT (2012a) Significance analysis and statistical dissection of variably methylated regions. Biostatistics 13:166–178
Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA (2012b) Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41:200–209
Jardim MJ (2011) microRNAs: implications for air pollution research. Mutat Res 717:38–45
Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreno L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E (2010) Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res 20:170–179
Jiang G, Xu L, Song S, Zhu C, Wu Q, Zhang L, Wu L (2008) Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 244:49–55
Jones B, Chen J (2006) Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J 25:2443–2452
Kakiuchi C, Ishiwata M, Nanko S, Ozaki N, Iwata N, Umekage T, Tochigi M, Kohda K, Sasaki T, Imamura A, Okazaki Y, Kato T (2008) Up-regulation of ADM and SEPX1 in the lymphoblastoid cells of patients in monozygotic twins discordant for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 147B:557–564
Kalfa N, Paris F, Soyer-Gobillard MO, Daures JP, Sultan C (2011) Prevalence of hypospadias in grandsons of women exposed to diethylstilbestrol during pregnancy: a multigenerational national cohort study. Fertil Steril 95:2574–2577
Kaminsky Z, Petronis A, Wang SC, Levine B, Ghaffar O, Floden D, Feinstein A (2008) Epigenetics of personality traits: an illustrative study of identical twins discordant for risk-taking behavior. Twin Res Hum Genet 11:1–11
Kanwal R, Gupta S (2011) Epigenetic modifications in cancer. Clin Genet 81:303–311
Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, Morris M, Haghighi F, Tycko B (2008) Genomic surveys by methylation-sensitive SNP analysis identify sequence- dependent allele-specific DNA methylation. Nat Genet 40:904–908
Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I (2006) Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet 38:149–153
Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT (2001) p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res 61:3419–3424
Klip H, Verloop J, van Gool JD, Koster ME, Burger CW, van Leeuwen FE (2002) Hypospadias in sons of women exposed to diethylstilbestrol in utero: a cohort study. Lancet 359:1102–1107
Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T (2007) Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet 16:2542–2551
Koukoura O, Sifakis S, Soufla G, Zaravinos A, Apostolidou S, Jones A, Widschwendter M, Spandidos DA (2011a) Loss of imprinting and aberrant methylation of IGF2 in placentas from pregnancies complicated with fetal growth restriction. Int J Mol Med 28:481–487
Koukoura O, Sifakis S, Zaravinos A, Apostolidou S, Jones A, Hajiioannou J, Widschwendter M, Spandidos DA (2011b) Hypomethylation along with increased H19 expression in placentas from pregnancies complicated with fetal growth restriction. Placenta 32:51–57
Kristensen LS, Wojdacz TK, Thestrup BB, Wiuf C, Hager H, Hansen LL (2009) Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer 9:453
Kuratomi G, Iwamoto K, Bundo M, Kusumi I, Kato N, Iwata N, Ozaki N, Kato T (2008) Aberrant DNA methylation associated with bipolar disorder identified from discordant monozygotic twins. Mol Psychiatry 13:429–441
Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC (2008) DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol 28:139–146
Laird PW (2010) Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11:191–203
Langevin SM, Houseman EA, Christensen BC, Wiencke JK, Nelson HH, Karagas MR, Marsit CJ, Kelsey KT (2011) The influence of aging, environmental exposures and local sequence features on the variation of DNA methylation in blood. Epigenetics 6:908–919
Langevin SM, Koestler DC, Christensen BC, Butler RA, Wiencke JK, Nelson HH, Houseman EA, Marsit CJ, Kelsey KT (2012) Peripheral blood DNA methylation profiles are indicative of head and neck squamous cell carcinoma: an epigenome-wide association study. Epigenetics 7:291–299
Lee H, Jaffe AE, Feinberg JI, Tryggvadottir R, Brown S, Montano C, Aryee MJ, Irizarry RA, Herbstman J, Witter FR, Goldman LR, Feinberg AP, Fallin MD (2012) DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. Int J Epidemiol 41:188–199
Li YF, Langholz B, Salam MT, Gilliland FD (2005) Maternal and grandmaternal smoking patterns are associated with early childhood asthma. Chest 127:1232–1241
Li L, Wang L, Xu X, Lou H, Le F, Sheng J, Huang H, Jin F (2011) Genome-wide DNA methylation patterns in IVF-conceived mice and their progeny: a putative model for ART-conceived humans. Reprod Toxicol 32:98–105
Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schubeler D (2011) Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43:1091–1097
Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, Tsui WY, Kong CK, Brunner HG, van Kessel AG, Yuen ST, van Krieken JH, Leung SY, Hoogerbrugge N (2009) Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet 41:112–117
Lillycrop KA (2011) Effect of maternal diet on the epigenome: implications for human metabolic disease. Proc Nutr Soc 70:64–72
Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC (2005) Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 135:1382–1386
Lim U, Song M-A (2012) Dietary and lifestyle factors of DNA methylation. In: Dumitrescu RG, Verma M (eds) Cancer epigenetics: methods and protocols, vol 863. Springer Science & Bysubess Neduam LLC, Berlin
Limsui D, Vierkant RA, Tillmans LS, Wang AH, Weisenberger DJ, Laird PW, Lynch CF, Anderson KE, French AJ, Haile RW, Harnack LJ, Potter JD, Slager SL, Smyrk TC, Thibodeau SN, Cerhan JR, Limburg PJ (2010) Cigarette smoking and colorectal cancer risk by molecularly defined subtypes. J Natl Cancer Inst 102:1012–1022
Liu X, Nelson A, Wang X, Kanaji N, Kim M, Sato T, Nakanishi M, Li Y, Sun J, Michalski J, Patil A, Basma H, Rennard SI (2009) MicroRNA-146a modulates human bronchial epithelial cell survival in response to the cytokine-induced apoptosis. Biochem Biophys Res Commun 380:177–182
Lovat F, Valeri N, Croce CM (2011) MicroRNAs in the pathogenesis of cancer. Semin Oncol 38:724–733
Lumey LH (1992) Decreased birthweights in infants after maternal in utero exposure to the Dutch famine of 1944–1945. Paediatr Perinat Epidemiol 6:240–253
Lumey LH, Stein AD, Susser E (2011) Prenatal famine and adult health. Annu Rev Public Health 32:237–262
Madrigano J, Baccarelli A, Mittleman MA, Wright RO, Sparrow D, Vokonas PS, Tarantini L, Schwartz J (2011) Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ Health Perspect 119:977–982
Majumdar S, Chanda S, Ganguli B, Mazumder DN, Lahiri S, Dasgupta UB (2010) Arsenic exposure induces genomic hypermethylation. Environ Toxicol 25:315–318
Marques CJ, Costa P, Vaz B, Carvalho F, Fernandes S, Barros A, Sousa M (2008) Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol Hum Reprod 14:67–74
Marsit CJ, Karagas MR, Danaee H, Liu M, Andrew A, Schned A, Nelson HH, Kelsey KT (2006) Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis 27:112–116
Mastroeni D, McKee A, Grover A, Rogers J, Coleman PD (2009) Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One 4:e6617
Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J, Mowry B, Hayward N (2007) Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol Psychiatry 12:815–825
Mattes J, Collison A, Plank M, Phipps S, Foster PS (2009) Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 106:18704–18709
McDaniell R, Lee BK, Song L, Liu Z, Boyle AP, Erdos MR, Scott LJ, Morken MA, Kucera KS, Battenhouse A, Keefe D, Collins FS, Willard HF, Lieb JD, Furey TS, Crawford GE, Iyer VR, Birney E (2010) Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328:235–239
McDonald SD, Han Z, Mulla S, Murphy KE, Beyene J, Ohlsson A (2009) Preterm birth and low birth weight among in vitro fertilization singletons: a systematic review and meta-analyses. Eur J Obstet Gynecol Reprod Biol 146:138–148
McDonald SD, Han Z, Mulla S, Ohlsson A, Beyene J, Murphy KE (2010) Preterm birth and low birth weight among in vitro fertilization twins: a systematic review and meta-analyses. Eur J Obstet Gynecol Reprod Biol 148:105–113
McGrath J, Solter D (1984) Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37:179–183
McMinn J, Wei M, Schupf N, Cusmai J, Johnson EB, Smith AC, Weksberg R, Thaker HM, Tycko B (2006) Unbalanced placental expression of imprinted genes in human intrauterine growth restriction. Placenta 27:540–549
Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, Wang SC, Petronis A (2008) Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet 82:696–711
Mitchell LE, Weinberg CR (2005) Evaluation of offspring and maternal genetic effects on disease risk using a family-based approach: the “pent” design. Am J Epidemiol 162:676–685
Mitchell MM, Lleo A, Zammataro L, Mayo MJ, Invernizzi P, Bach N, Shimoda S, Gordon S, Podda M, Gershwin ME, Selmi C, LaSalle JM (2011) Epigenetic investigation of variably X chromosome inactivated genes in monozygotic female twins discordant for primary biliary cirrhosis. Epigenetics 6:95–102
Molitor J, Marjoram P, Thomas DC (2003) Fine-scale mapping of diseases with multiple mutations via spatial clustering techniques. Am J Hum Genet 73:1368–1384
Moller H, Jorgensen N, Forman D (1995) Trends in incidence of testicular cancer in boys and adolescent men. Int J Cancer 61:761–764
Morgan HD, Sutherland HG, Martin DI, Whitelaw E (1999) Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23:314–318
Murphy SK, Adigun A, Huang Z, Overcash F, Wang F, Jirtle RL, Schildkraut JS, Murtha AP, Iversen ES, Hoyo C (2011) Gender-specific methylation differences in relation to prenatal exposure to cigarette smoke. Gene 494:36–43
Nadeau K, McDonald-Hyman C, Noth EM, Pratt B, Hammond SK, Balmes J, Tager I (2010) Ambient air pollution impairs regulatory T-cell function in asthma. J Allergy Clin Immunol 126:845–852
Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM (2006) Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics 1:e1–e11
Nan HM, Song YJ, Yun HY, Park JS, Kim H (2005) Effects of dietary intake and genetic factors on hypermethylation of the hMLH1 gene promoter in gastric cancer. World J Gastroenterol 11:3834–3841
Nana-Sinkam SP, Hunter MG, Nuovo GJ, Schmittgen TD, Gelinas R, Galas D, Marsh CB (2009) Integrating the MicroRNome into the study of lung disease. Am J Respir Crit Care Med 179:4–10
Nelissen EC, van Montfoort AP, Dumoulin JC, Evers JL (2011) Epigenetics and the placenta. Hum Reprod Update 17:397–417
Nguyen A, Rauch TA, Pfeifer GP, Hu VW (2010) Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB J 24:3036–3051
Oates NA, van Vliet J, Duffy DL, Kroes HY, Martin NG, Boomsma DI, Campbell M, Coulthard MG, Whitelaw E, Chong S (2006) Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet 79:155–162
O’Hanlon TP, Rider LG, Gan L, Fannin R, Paules RS, Umbach DM, Weinberg CR, Shah RR, Mav D, Gourley MF, Miller FW (2011) Gene expression profiles from discordant monozygotic twins suggest that molecular pathways are shared among multiple systemic autoimmune diseases. Arthr Res Ther 13:R69
Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, Berman DM, Jenuwein T, Pruitt K, Sharkis SJ, Watkins DN, Herman JG, Baylin SB (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet 39:237–242
Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, Abdul Aziz NK, Carlin JB, Morley R, Saffery R, Craig JM (2010) DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 19:4176–4188
Pacheco SE, Houseman EA, Christensen BC, Marsit CJ, Kelsey KT, Sigman M, Boekelheide K (2011) Integrative DNA methylation and gene expression analyses identify DNA packaging and epigenetic regulatory genes associated with low motility sperm. PLoS One 6:e20280
Palmer JR, Wise LA, Robboy SJ, Titus-Ernstoff L, Noller KL, Herbst AL, Troisi R, Hoover RN (2005) Hypospadias in sons of women exposed to diethylstilbestrol in utero. Epidemiology 16:583–586
Patel CJ, Bhattacharya J, Butte AJ (2010) An environment-wide association study (EWAS) on type 2 diabetes mellitus. PLoS One 5:e10746
Pauley KM, Chan EK (2008) MicroRNAs and their emerging roles in immunology. Ann N Y Acad Sci 1143:226–239
Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, Baccarelli A (2009) Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer 125:1692–1697
Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J (2006) Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14:159–166
Perera F, Herbstman J (2011) Prenatal environmental exposures, epigenetics, and disease. Reprod Toxicol 31:363–373
Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, Ho SM (2009) Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One 4:E4488
Pilsner JR, Hu H, Ettinger A, Sanchez BN, Wright RO, Cantonwine D, Lazarus A, Lamadrid-Figueroa H, Mercado-Garcia A, Tellez-Rojo MM, Hernandez-Avila M (2009) Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect 117:1466–1471
Pirola L, Balcerczyk A, Tothill RW, Haviv I, Kaspi A, Lunke S, Ziemann M, Karagiannis T, Tonna S, Kowalczyk A, Beresford-Smith B, Macintyre G, Kelong M, Hongyu Z, Zhu J, El-Osta A (2011) Genome-wide analysis distinguishes hyperglycemia regulated epigenetic signatures of primary vascular cells. Genome Res 21:1601–1615
Plagemann A, Harder T, Brunn M, Harder A, Roepke K, Wittrock-Staar M, Ziska T, Schellong K, Rodekamp E, Melchior K, Dudenhausen JW (2009) Hypothalamic proopiomelanocortin promoter methylation becomes altered by early overfeeding: an epigenetic model of obesity and the metabolic syndrome. J Physiol 587:4963–4976
Pons JC, Papiernik E, Billon A, Hessabi M, Duyme M (2005) Hypospadias in sons of women exposed to diethylstilbestrol in utero. Prenat Diagn 25:418–419
Pozharny Y, Lambertini L, Ma Y, Ferrara L, Litton CG, Diplas A, Jacobs AR, Chen J, Stone JL, Wetmur J, Lee MJ (2010) Genomic loss of imprinting in first-trimester human placenta. Am J Obstet Gynecol 202(391):e1–e8
Ptashne M (2007) On the use of the word ‘epigenetic’. Curr Biol 17:R233–R236
Rakyan VK, Down TA, Balding DJ, Beck S (2011) Epigenome-wide association studies for common human diseases. Nat Rev Genet 12:529–541
Reddy MA, Natarajan R (2011) Epigenetic mechanisms in diabetic vascular complications. Cardiovasc Res 90:421–429
Relton CL, Davey Smith G (2012) Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol 41:161–176
Rosa A, Picchioni MM, Kalidindi S, Loat CS, Knight J, Toulopoulou T, Vonk R, van der Schot AC, Nolen W, Kahn RS, McGuffin P, Murray RM, Craig IW (2008) Differential methylation of the X-chromosome is a possible source of discordance for bipolar disorder female monozygotic twins. Am J Med Genet B Neuropsychiatr Genet 147B:459–462
Russo AL, Thiagalingam A, Pan H, Califano J, Cheng KH, Ponte JF, Chinnappan D, Nemani P, Sidransky D, Thiagalingam S (2005) Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res 11:2466–2470
Saffery R, Morley R, Carlin JB, Joo J-HE, Ollikainen M, Novakovic B, Andronikos R, Li X, Loke YJ, Carson N, Wallace EM, Umstad MP, Permezel M, Galati JC, Craig JM (2011) Cohort profile: the peri/post-natal epigenetic twins study. Int J Epidemiol 41:55–61
Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD (2012) Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol 129(232–239):e7
Samowitz WS, Albertsen H, Sweeney C, Herrick J, Caan BJ, Anderson KE, Wolff RK, Slattery ML (2006) Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst 98:1731–1738
Sato A, Otsu E, Negishi H, Utsunomiya T, Arima T (2007) Aberrant DNA methylation of imprinted loci in superovulated oocytes. Hum Reprod 22:26–35
Savage T, Peek J, Hofman PL, Cutfield WS (2011) Childhood outcomes of assisted reproductive technology. Hum Reprod 26:2392–2400
Scesnaite A, Jarmalaite S, Mutanen P, Anttila S, Nyberg F, Benhamou S, Boffetta P, Husgafvel-Pursiainen K (2012) Similar DNA methylation pattern in lung tumours from smokers and never-smokers with second-hand tobacco smoke exposure. Mutagenesis [Epub ahead of print]
Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J (2010) Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet 86:196–212
Schernhammer ES, Giovannuccci E, Fuchs CS, Ogino S (2008) A prospective study of dietary folate and vitamin B and colon cancer according to microsatellite instability and KRAS mutational status. Cancer Epidemiol Biomarkers Prev 17:2895–2898
Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H (2007) Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet 39:232–236
Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, Frauenknecht K, Riepert T, Tresch A, Muller AM, Coerdt W, Zechner U, Haaf T (2010) Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res 38:3880–3890
Schroeder DI, Lott P, Korf I, LaSalle JM (2011) Large-scale methylation domains mark a functional subset of neuronally expressed genes. Genome Res 21:1583–1591
Serman L, Vlahovic M, Sijan M, Bulic-Jakus F, Serman A, Sincic N, Matijevic R, Juric-Lekic G, Katusic A (2007) The impact of 5-azacytidine on placental weight, glycoprotein pattern and proliferating cell nuclear antigen expression in rat placenta. Placenta 28:803–811
Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31:27–36
Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, Montgomery SB, Dupre Y, Antonarakis SE (2011) DNA methylation profiles of human active and inactive X chromosomes. Genome Res 21:1592–1600
Siebel AL, Fernandez AZ, El-Osta A (2010) Glycemic memory associated epigenetic changes. Biochem Pharmacol 80:1853–1859
Siegmund KD (2011) Statistical approaches for the analysis of DNA methylation microarray data. Hum Genet 129:585–595
Siegmund KD, Laird PW, Laird-Offringa IA (2004) A comparison of cluster analysis methods using DNA methylation data. Bioinformatics 20:1896–1904
Siegmund KD, Levine AJ, Chang J, Laird PW (2006) Modeling exposures for DNA methylation profiles. Cancer Epidemiol Biomarkers Prev 15:567–572
Sinclair KD, Young LE, Wilmut I, McEvoy TG (2000) In utero overgrowth in ruminants following embryo culture: lessons from mice and a warning to men. Hum Reprod 15(Suppl 5):68–86
Slatkin M (2009) Epigenetic inheritance and the missing heritability problem. Genetics 182:845–850
Slattery ML, Anderson K, Curtin K, Ma KN, Schaffer D, Samowitz W (2001) Dietary intake and microsatellite instability in colon tumors. Int J Cancer 93:601–607
Slattery ML, Curtin K, Sweeney C, Levin TR, Potter JD, Wolff RK, Albertsen H, Samowitz W (2006) Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer 120:656–663
Su RC, Becker AB, Kozyrskyj AL, Hayglass KT (2008) Epigenetic regulation of established human type 1 versus type 2 cytokine responses. J Allergy Clin Immunol 121(57–63):e3
Sundar IK, Caito S, Yao H, Rahman I (2010) Oxidative stress, thiol redox signaling methods in epigenetics. Methods Enzymol 474:213–244
Suter CM, Martin DI, Ward RL (2004) Germline epimutation of MLH1 in individuals with multiple cancers. Nat Genet 36:497–501
Swan SH (2000) Intrauterine exposure to diethylstilbestrol: long-term effects in humans. APMIS 108:793–804
Tal O, Kisdi E, Jablonka E (2010) Epigenetic contribution to covariance between relatives. Genetics 184:1037–1050
Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, Putter H, Slagboom PE, Heijmans BT (2010) Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J 24:3135–3144
Talens RP, Jukema JW, Trompet S, Kremer D, Westendorp RG, Lumey LH, Sattar N, Putter H, Slagboom PE, Heijmans BT (2011) Hypermethylation at loci sensitive to the prenatal environment is associated with increased incidence of myocardial infarction. Int J Epidemiol 40:1–10
Tanay A, O’Donnell AH, Damelin M, Bestor TH (2007) Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci USA 104:5521–5526
Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, Bertazzi PA, Baccarelli A (2009) Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect 117:217–222
Tarutani Y, Takayama S (2011) Monoallelic gene expression and its mechanisms. Current opinion in plant biology 14:608–613
Taylor PD, Poston L (2007) Developmental programming of obesity in mammals. Exp Physiol 92:287–298
Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM (2011) DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics 6:828–837
Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, Kocjan G, Gayther SA, Jones A, Beck S, Wagner W, Laird PW, Jacobs IJ, Widschwendter M (2010) Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res 20:440–446
Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A (2010) CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464:1082–1086
Tierling S, Souren NY, Reither S, Zang KD, Meng-Hentschel J, Leitner D, Oehl-Jaschkowitz B, Walter J (2011) DNA methylation studies on imprinted loci in a male monozygotic twin pair discordant for Beckwith-Wiedemann syndrome. Clin Genet 79:546–553
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Nat Acad Sci USA 96:8681–8686
Trasler JM (2009) Epigenetics in spermatogenesis. Mol Cell Endocrinol 306:33–36
Tycko B (2010) Mapping allele-specific DNA methylation: a new tool for maximizing information from GWAS. Am J Hum Genet 86:109–112
Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res 32:4100–4108
van den Berg BJ, Christianson RE, Oechsli FW (1988) The California Child Health and Development Studies of the School of Public Health, University of California at Berkeley. Paediatr Perinat Epidemiol 2:265–282
van den Donk M, van Engeland M, Pellis L, Witteman BJ, Kok FJ, Keijer J, Kampman E (2007) Dietary folate intake in combination with MTHFR C677T genotype and promoter methylation of tumor suppressor and DNA repair genes in sporadic colorectal adenomas. Cancer Epidemiol Biomarkers Prev 16:327–333
Van Guelpen B, Dahlin AM, Hultdin J, Eklof V, Johansson I, Henriksson ML, Cullman I, Hallmans G, Palmqvist R (2010) One-carbon metabolism and CpG island methylator phenotype status in incident colorectal cancer: a nested case-referent study. Cancer Causes Control 21:557–566
van Montfoort AP, Hanssen LL, de Sutter P, Viville S, Geraedts JP, de Boer P (2012) Assisted reproduction treatment and epigenetic inheritance. Hum Reprod Update 18:171–197
Vermeulen SH, Shi M, Weinberg CR, Umbach DM (2009) A hybrid design: case-parent triads supplemented by control-mother dyads. Genet Epidemiol 33:136–144
Vessey MP, Fairweather DV, Norman-Smith B, Buckley J (1983) A randomized double-blind controlled trial of the value of stilboestrol therapy in pregnancy: long-term follow-up of mothers and their offspring. Br J Obstet Gynaecol 90:1007–1017
Wark PA, Weijenberg MP, van’t Veer P, van Wijhe G, Luchtenborg M, van Muijen GN, de Goeij AF, Goldbohm RA, van den Brandt PA (2005) Fruits, vegetables, and hMLH1 protein-deficient and -proficient colon cancer: the Netherlands cohort study. Cancer Epidemiol Biomarkers Prev 14:1619–1625
Waterland RA, Jirtle RL (2003) Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 23:5293–5300
Waterland RA, Jirtle RL (2004) Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition 20:63–68
Waterland RA, Kellermayer R, Laritsky E, Rayco-Solon P, Harris RA, Travisano M, Zhang W, Torskaya MS, Zhang J, Shen L, Manary MJ, Prentice AM (2010) Season of conception in rural gambia affects DNA methylation at putative human metastable epialleles. PLoS Genet 6:e1001252
Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ (2004) Epigenetic programming by maternal behavior. Nat Neurosci 7:847–854
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38:787–793
Weth O, Renkawitz R (2011) CTCF function is modulated by neighboring DNA binding factors. Biochem Cell Biol 89:459–468
White GP, Watt PM, Holt BJ, Holt PG (2002) Differential patterns of methylation of the IFN-gamma promoter at CpG and non-CpG sites underlie differences in IFN-gamma gene expression between human neonatal and adult CD45RO-T cells. J Immunol 168:2820–2827
Whitehead ED, Leiter E (1981) Genital abnormalities and abnormal semen analyses in male patients exposed to diethylstilbestrol in utero. J Urol 125:47–50
Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW (2007) Epigenetic stem cell signature in cancer. Nat Genet 39(2):157–158
Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ (2010) Epigenetics in atherosclerosis and inflammation. J Cell Mol Med 14:1225–1240
Wilker EH, Baccarelli A, Suh H, Vokonas P, Wright RO, Schwartz J (2010) Black carbon exposures, blood pressure, and interactions with single nucleotide polymorphisms in MicroRNA processing genes. Environ Health Perspect 118:943–948
Wong CC, Caspi A, Williams B, Craig IW, Houts R, Ambler A, Moffitt TE, Mill J (2010) A longitudinal study of epigenetic variation in twins. Epigenetics 5:516–526
Wong CC, Caspi A, Williams B, Houts R, Craig IW, Mill J (2011) A longitudinal twin study of skewed X chromosome-inactivation. PLoS One 6:e17873
Wright RO, Baccarelli A (2007) Metals and neurotoxicology. J Nutr 137:2809–2813
Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A (2010) Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 118:790–795
Wu HC, Delgado-Cruzata L, Flom JD, Kappil M, Ferris JS, Liao Y, Santella RM, Terry MB (2011) Global methylation profiles in DNA from different blood cell types. Epigenetics 6:76–85
Yauk CL, Berndt ML, Williams A, Rowan-Carroll A, Douglas GR, Stampfli MR (2007) Mainstream tobacco smoke causes paternal germ-line DNA mutation. Cancer Res 67:5103–5106
Yauk C, Polyzos A, Rowan-Carroll A, Somers CM, Godschalk RW, Van Schooten FJ, Berndt ML, Pogribny IP, Koturbash I, Williams A, Douglas GR, Kovalchuk O (2008) Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Nat Acad Sci 105:605–610
Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, Yin Y, Wang C, Zhang T, Zhu D, Zhang D, Xu J, Chen Q, Ba Y, Liu J, Wang Q, Chen J, Wang J, Wang M, Zhang Q, Zhang J, Zen K, Zhang CY (2012) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res 22:107–126
Zhou ZH, Lei YX, Wang CX (2011) Analysis of aberrant methylation in DNA repair genes during malignant transformation of human bronchial epithelial cells induced by cadmium. Toxicol Sci 125:412–417
Acknowledgments
Supported in part by NIH grants R21 ES020794, P30 ES 07048, P30 CA014089 and a Whittier Foundation grant to the Norris Comprehensive Cancer Center.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
V. K. Cortessis and D. C. Thomas contributed equally to this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Cortessis, V.K., Thomas, D.C., Levine, A.J. et al. Environmental epigenetics: prospects for studying epigenetic mediation of exposure–response relationships. Hum Genet 131, 1565–1589 (2012). https://doi.org/10.1007/s00439-012-1189-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-012-1189-8