Abstract
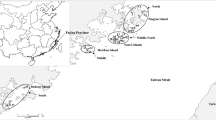
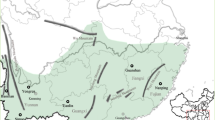
In this study, outcrossing rates and genetic diversity in natural populations of Nelumbo nucifera were investigated. The estimated multilocus outcrossing rate (tm) based on 28 ISSR loci was over 90%. Analysis of genetic diversity revealed that this index was high at the species level (Hs = 0.325, I = 0.514), but low within the individual study populations (Hs = 0.148, I = 0.212). Gst-B was 0.547 and Nm was 0.414. The results of AMOVA indicated that 54.6% of the variation was due to the difference between the regions and 45.4% to the variation within the region. Although the populations were predominantly outcrossing, most of the genetic diversity was attributed to geographical effects instead of their habitats because low sexual recruitment and clonal growth deeply reduced the genetic diversity within the populations. On the basis of the high tm, Gst-B and low Nm values, we recommended that any future conservation plans should include both in situ conservation and germplasm collection.


Similar content being viewed by others
References
Adams WT (1983) Applications of isozymes in tree breeding. In: Tanksley SD, Orton TJ (eds) Isozymes in plant genetics and breeding, Part A. Elsevier Science Publishers, Amsterdam, pp 60–64
Allard RW (1975) The mating system and microevolution. Genetics 79:115–126
Bornet B, Goraguer F, Joly G, Branchard M (2002) Genetic diversity in European and Argentinean cultivated potatoes (Solanum tuberosum subsp. tuberosum) detected by inter-simple sequence repeats (ISSRs). Genome 45:481–484
Bridle JR, Pedro PM, Butlin RK (2004) Habitat fragmentation and biodiversity: testing for the evolutionary effects of refugia. Evolution 58:1394–1396
Dong YC, Zheng DS (2005) Summary of national key agriculture wild plant protection. Chinese Meteorologic Publishing Company, Beijing, p 122
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distance among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics 131:479–491
Friedman WE, Moore RC, Purugganan MD (2004) The evolution of plant development. Am J Bot 91:1726–1741
Fritsch P, Rieseberg LH (1992) High outcrossing rates maintain male and hermaphrodite individuals in populations of the flowering plant Datisca glomerata. Nature 359:633–636
Gaiotto FA, Bramucci M, Grattapaglia D (1997) Estimation of outcrossing rate in a breeding population of Eucalyptus urophylla with dominant RAPD and AFLP markers. Theor Appl Genet 95:842–849
Ge XJ, Sun M (1999) Reproductive biology and genetic diversity of a crypto viviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and intersimple sequence repeat (ISSR) analysis. Mol Ecol 8:2061–2069
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer, Sunderland, pp 43–63
Han YC, Teng CZ, Zhong S, Zhou MQ, Hu ZL, Song YC (2007) Genetic variation and clonal diversity in populations of Nelumbo nucifera (Nelumbonaceae) in Central China detected by ISSR markers. Aquat Bot 86:69–75
Holsinger K (1999) Analysis of genetic diversity in geographically structured populations: a Bayesian perspective. Hereditas 130:245–255
Holsinger KE, Lewis PO (2003) HICKORY: a package for analysis of population genetic data, version 1.0. Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs
Holsinger KE, Wallace LE (2004) Bayesian approaches for the analysis of population genetic structure: an example from Platanthera leucophaea (Orchidaceae). Mol Ecol 13:887–894
Holsinger KE, Lewis PO, Dey DK (2002) A Bayesian approach to inferring population structure from dominant markers. Mol Ecol 11:1157–1164
Kojima T, Nagaoka T, Noda K, Ogihara Y (1998) Genetic linkage map of ISSR and RAPD markers in einkorn wheat in relation to that of RFLP markers. Theor Appl Genet 96:37–45
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mantel NA (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Miller MP (1998) AMOVA-PREP 1.01: a program for the preparation of the AMOVA input files from dominant-marker raw data. Department of Biological Sciences, Northern Arizona University, Flagstaff, AZ
Mukherjee PK, Saha K, Das J, Pal M, Saha BP (1997) Studies on the anti-inflammatory activity of rhizomes of Nelumbo nucifera. Planta Med 63:367–369
Muona O (1990) Population genetics in forest tree improvement. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics, breeding and genetic resources. Sinauer Press, Sunderland, MA, USA, pp 282–298
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3(2):93–114
Qian JQ (2002) Cardiovascular pharmacological effects of bisbenzylisoquinoline alkaloid derivatives. Acta Pharmacol Sin 23:1086–1092
Real LA (1994) Ecological genetics. Princeton University Press, Princeton, NJ
Richards CM, Church S, McCauley DE (1999) The influence of population size and isolation on gene flow by pollen in Silene alba. Evolution 53:63–73
Ritland K (2004) Multilocus mating system program MLTR. http://genetics.forestry.ubc.ca/ritland/programs
Ritland K, Jain SA (1981) Model for the estimation of outcrossing rate and gene frequencies using ‘‘n’’ independent loci. Heredity 47:35–52
Rohlf FJ (1992) NTSYS–PC: numerical taxonomy and multivariate analysis system, version 2.0. State University of New York, Stony Brook, NY
Schaal BA, Hayworth DA, Olsen KM, Rauscher JT, Smith WA (1998) Phylogeographic studies in plants: problems and prospects. Mol Ecol 7:465–474
Schaal BA, Leverich WJ, Rogstad SH (1991) Comparison of methods for assessing genetic variation in plant conservation biology. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 123–134
Sinha S, Mukherjee PK, Mukherjee K, Pal M, Mandal SC, Saha BP (2000) Evaluation of antipyretic potential of Nelumbo nucifera stalk extract. Phytother Res 14:272–274
Tigerstedt PMA (1984) Genetic mechanisms for adaptation: the mating system of Scots pine. In: Swaminathan MS, Chopra VL, Joshi BC, Sharma RP, Bansal HC (eds) Genetics: new frontiers (Proc 15th Int Congr Genetics). Oxford and IBH Publ, New Delhi, pp 317–322
Wang QC, Zhang YX (2005) Lotus varieties of China. Chinese Forestry Publishing Company, Beijing
Whitlock MC, McCauley DE (1990) Some population genetic consequences of colony formation and extinction: genetic correlations within founding groups. Evolution 44:1717–1724
Xue JH, Zhou LH, Zhou SL (2006) Genetic diversity and geographic pattern of wild lotus (Nelumbo nucifera) in Heilongjiang Province. Chin Sci Bull 51:421–432
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183
Acknowledgments
We thank Prof. Wang QF and Dr. Chen JM (of the School of Life Sciences, Wuhan University), Prof. Ge S and Dr. Zhu QH (of the Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences) for their help with data analysis. This work was supported by the National Natural Science Foundation of China (No. 30771375 and No. 30471114) and National Science and Technology Pillar Program (No. 2007BAD37B06).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Han, YC., Teng, CZ., Wahiti, G.R. et al. Mating system and genetic diversity in natural populations of Nelumbo nucifera (Nelumbonaceae) detected by ISSR markers. Plant Syst Evol 277, 13–20 (2009). https://doi.org/10.1007/s00606-008-0096-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00606-008-0096-x