Abstract
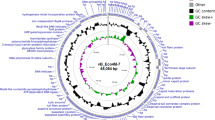
Poultry intestinal material, sewage and poultry processing drainage water were screened for virulent Clostridium perfringens bacteriophages. Viruses isolated from broiler chicken offal washes (O) and poultry feces (F), designated ΦCP39O and ΦCP26F, respectively, produced clear plaques on host strains. Both bacteriophages had isometric heads of 57 nm in diameter with 100-nm non-contractile tails characteristic of members of the family Siphoviridae in the order Caudovirales. The double-strand DNA genome of bacteriophage ΦCP39O was 38,753 base pairs (bp), while the ΦCP26F genome was 39,188 bp, with an average GC content of 30.3%. Both viral genomes contained 62 potential open reading frames (ORFs) predicted to be encoded on one strand. Among the ORFs, 29 predicted proteins had no known similarity while others encoded putative bacteriophage capsid components such as a pre-neck/appendage, tail, tape measure and portal proteins. Other genes encoded a predicted DNA primase, single-strand DNA-binding protein, terminase, thymidylate synthase and a transcription factor. Potential lytic enzymes such as a fibronectin-binding autolysin, an amidase/hydrolase and a holin were encoded in the viral genomes. Several ORFs encoded proteins that gave BLASTP matches with proteins from Clostridium spp. and other Gram-positive bacterial and bacteriophage genomes as well as unknown putative Collinsella aerofaciens proteins. Proteomics analysis of the purified viruses resulted in the identification of the putative pre-neck/appendage protein and a minor structural protein encoded by large open reading frames. Variants of the portal protein were identified, and several mycobacteriophage gp6-like protein variants were detected in large amounts relative to other virion proteins. The predicted amino acid sequences of the pre-neck/appendage proteins had major differences in the central portion of the protein between the two phage gene products. Based on phylogenetic analysis of the large terminase protein, these phages are predicted to be pac-type, using a head-full DNA packaging strategy.




Similar content being viewed by others
References
Ackermann H-W (1998) Tailed bacteriophages: the order Caudovirales. Adv Virus Res 51:135–201
Ackermann H-W (2003) Bacteriophage observations and evolution. Res Microbiol 154:245–251
Ackermann H-W (2008) Basic phage electron microscopy. Methods Mol Biol 501:113–126
Aebersold RH, Leavitt J, Saavedra RA, Hood LE, Kent SB (1987) Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion on nitrocellulose. Proc Natl Acad Sci USA 84:6970–6974
Alonso JC, Lüder G, Stiege AC, Chai S, Weise F, Trautner TA (1997) The complete nucleotide sequence and functional organization of Bacillus subtilis bacteriophage SPP1. Gene 204:201–212
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Becker B, De la Fuente N, Gassel M, Günther D, Tavares P, Lurz R, Trautner TA, Alonso JC (1997) Head morphogenesis genes of the Bacillus subtilis bacteriophage SPP1. J Mol Biol 268:822–839
Bernhardt TG, Wang IN, Struck DK, Young R (2002) Breaking free: “protein antibiotics” and phage lysis. Res Microbiol 153:493–501
Bettegowda C, Huang X, Lin J, Cheong I, Kohli M, Szabo SA, Zhang X, Diaz LA Jr, Velculescu VE, Parmigiani G, Kinzler KW, Vogelstein B, Zhou S (2006) The genome and transcriptomes of the anti-tumor agent Clostridium novyi-NT. Nat Biotechnol 24:1573–1580
Boyd EF, Brüssow H (2002) Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol 10:521–529
Bradley DE, Hoeniger JF (1971) Structural changes in cells of Clostridium perfringens infected with a short-tailed bacteriophage. Can J Microbiol 17:397–402
Brüggemann H (2005) Genomics of clostridial pathogens: implication of extrachromosomal elements in pathogenicity. Curr Opin Microbiol 8:601–605
Brüggemann H, Baumer S, Fricke WF, Wiezer A, Liesegang H, Decker I, Herzberg C, Martinez-Arias R, Merkl R, Henne A, Gottschalk G (2003) The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc Natl Acad Sci USA 100:1316–1321
Brüssow H, Canchaya C, Hardt WD (2004) Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev 68:560–602
Brussow H, Desiere F (2001) Comparative phage genomics and the evolution of Siphoviridae: insights from dairy phages. Mol Microbiol 39:213–222
Burrus V, Pavlovic G, Decaris B, Guedon G (2002) The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97
Camacho AG, Gual A, Lurz R, Tavares P, JAlonso JC (2003) Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem 278:23251–23259
Conard B, Cole ST (1990) Lysogenic phages of Clostridium perfringens: mapping of the chromosomal attachment sites. FEMS Microbiol Lett 54:323–326
Casjens SR (2005) Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol 8:451–458
Casjens SR, Gilcrease EB (2009) Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol Biol 502:91–111
Champion KM, Nishihara JC, Joly JC, Arnott D (2001) Similarity of the Escherichia coli proteome upon completion of different biopharmaceutical fermentation processes. Proteomics 1:1133–1148
Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JA (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44:812–826
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Eklund MW, Poysky FT, Reed SM, Smith CA (1971) Bacteriophage and the toxigenicity of Clostridium botulinum type C. Science 172:480–482
Eklund MW, Poysky FT, Reed SM (1972) Bacteriophage and the toxigenicity of Clostridium botulinum type D. Nat New Biol 235:16–17
Fane BA, Prevelige PE Jr (2003) Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem 64:259–299
Fischetti VA (2005) Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol 13:491–496
Fouts DE, Rasko DA, Cer RZ, Jiang L, Fedorova NB, Shvartsbeyn A, Vamathevan JJ, Tallon L, Althoff R, Arbogast TS, Fadrosh DW, Read TD, Gill SR (2006) Sequencing Bacillus anthracis typing phages Gamma and Cherry reveals a common ancestry. J Bacteriol 188:3402–3408
Finhout E, Lee K (2003) Comparison of automated in-gel digest methods for femtomole level samples. Electrophoresis 24:3508–3516
Garwes DJ, Pike BV, Wyld SG, Pocock DH, Gourlay RN (1975) Characterization of Mycoplasmatales virus-laidlawii 3. J Gen Virol 29:11–24
Goh S, Riley TV, Chang BJ (2005) Isolation and characterization of temperate bacteriophages of Clostridium difficile. Appl Environ Microbiol 71:1079–1083
Goh S, Ong PF, Song KP, Riley TV, Chang BJ (2007) The complete genome sequence of Clostridium difficile phage ΦC2 and comparisons to ΦCD119 and inducible prophages of CD630. Microbiology 153:676–685
Gorg A, Weiss W, Dunn M (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665–3685
Govind R, Fralick JA, Rolfe RD (2006) Genomic organization and molecular characterization of Clostridium difficile bacteriophage ΦCD119. J Bacteriol 188:2568–2577
Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA (2009) Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol 83:12037–12045
Grant RB, Riemann HP (1976) Temperate phages of Clostridium perfringens type C1. Can J Microbiol 22:603–610
Helms C, Graham MY, Dutchik JE, Olson MV (1985) A new method for purifying lambda DNA from phage lysates. DNA 4:39–49
Hermoso JA, Monterroso B, Albert A, Galán B, Ahrazem O, García P, Martínez-Ripoll M, García JL, Menéndez M (2003) Structural basis for selective recognition of pneumococcal cell wall by modular endolysin from phage Cp-1. Structure 11:1239–1249
Hirano S, Yonekura Y (1967) The structure of Clostridium perfringens bacteriophages. Acta Med Univ Kagoshima 9:41–56
Kwan T, Liu J, DuBow M, Gros P, Pelletier J (2005) The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc Natl Acad Sci USA 102:5174–5179
Lahm HW, Langen H (2000) Mass spectrometry: a tool for the identification of proteins separated by gels. Electrophoresis 21:2105–2114
Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE (2006) The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 312:1791–1795
Lewis JG, Chang G-J, Lanciotti RS, Trent DW (1992) Direct sequencing of large flavivirus PCR products for analysis of genome variation and molecular epidemiological investigations. J Virol Meth 38:11–24
Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, McCarty J, Srikumar R, Williams D, Wu JJ, Gros P, Pelletier J, DuBow M (2004) Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol 22:185–191
Liu X, Kong S, Shi M, Fu L, GaoY AnC (2008) Genomic analysis of freshwater cyanophage Pf-WMP3 Infecting cyanobacterium Phormidium foveolarum: the conserved elements for a phage. Microb Ecol 56:671–680
Loessner MJ (2005) Bacteriophage endolysins—current state of research and applications. Curr Opin Microbiol 8:480–487
Lucchini S, Desiere F, Brüssow H (1998) The structural gene module in Streptococcus thermophilus bacteriophage ΦSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology 246:63–73
Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26:1107–1115
Mahony DE, Easterbrook KB (1970) Intracellular development of a bacteriophage of Clostridium perfringens. Can J Microbiol 16:983–988
Mahony DE, Kalz GG (1968) A temperate bacteriophage of Clostridium perfringens. Can J Microbiol 14:1085–1093
Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35:D237–D240
Margulies M, Egholm M, Altman WE, Attiya S, JBader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Mayer MJ, Narbad A, Gasson MJ (2008) Molecular characterization of a Clostridium difficile bacteriophage and its cloned biologically active endolysin. J Bacteriol 190:6734–6740
McCrea JF, Epstein RS, Barry WH (1961) Use of potassium tartrate for equilibrium density-gradient centrifugation of animal viruses. Nature 189:220–221
Mead DA, Pey NK, Herrnstadt C, Marcil RA, Smith LM (1991) A universal method for the direct cloning of PCR amplified nucleic acid. Biotechnology 9:657–662
Merril CR, Scholl D, Adhya SL (2003) The prospect for bacteriophage therapy in Western medicine. Nat Rev Drug Discov 2:489–497
Misra V, Blumenthal RM, Babiuk LA (1981) Proteins specified by bovine herpesvirus 1 (infectious bovine rhinotracheitis virus). J Virol 40:367–378
Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, Anson EL, Bolanos RA, Chou HH, Jordan CM, Halpern AL, Lonardi S, Beasley EM, Brandon RC, Chen L, Dunn PJ, Lai Z, Liang Y, Nusskern DR, Zhan M, Zhang Q, Zheng X, Rubin GM, Adams MD, Venter JC (2000) A whole-genome assembly of Drosophila. Science 287:2196–2204
Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan SJ, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT (2006) Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res 16:1031–1040
O’Farrell PH (1975) High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021
Paquette G, Fredette V (1977) Properties of four temperate bacteriophages active on Clostridium perfringens type A. Rev Can Biol 36:205–215
Paredes CJ, Alsaker KV, Papoutsakis ET (2005) A comparative genomic view of clostridial sporulation and physiology. Nat Rev Microbiol 3:969–978
Paul JH, Sullivan MB, Segall AM, Rohwer F (2002) Marine phage genomics. Comp Biochem Physiol B Biochem Mol Biol 133:463–476
Payment P, Franco E (1993) Clostridium perfringens and somatic coliphages as indicators of the efficiency of drinking water treatment for viruses and protozoan cysts. Appl Environ Microbiol 59:2418–2424
Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, Brucker W, Kumar V, Kandasamy J, Keenan L, Bardarov S, Kriakov J, Lawrence JG, Jacobs WR Jr, Hendrix RW, Hatfull GF (2003) Origins of highly mosaic mycobacteriophage genomes. Cell 113:171–182
Pommier S, Gavioli M, Cascales E, Lloubès R (2005) Tol-dependent macromolecule import through the Escherichia coli cell envelope requires the presence of an exposed TolA binding motif. J Bacteriol 187:7526–7534
Sambrook J, Maniatis T, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Sawires YS, Songer JG (2006) Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe 12:23–43
Schäffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF (2001) Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res 29:2994–3005
Schmidt HA, von Haeseler A (2007) Maximum-likelihood analysis using TREE-PUZZLE. Curr Protoc Bioinformatics, Chapter 6: Unit 6.6
Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J (2006) The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786
Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H (2002) Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc Natl Acad Sci USA 99:996–1001
Siragusa GR, Danyluk MD, Hiett KL, Wise MG, Craven SE (2006) Molecular subtyping of poultry-associated type A Clostridium perfringens isolates by repetitive-element PCR. J Clin Microbiol 44:1065–1073
Smedley JG 3rd, Fisher DJ, Sayeed S, Chakrabarti G, McClane BA (2004) The enteric toxins of Clostridium perfringens. Rev Physiol Biochem Pharmacol 152:183–204
Smith HW (1959) The bacteriophages of Clostridium perfringens. J Gen Microbiol 21:622–630
Smith LM, Sanders JZ, Kaiser RJ, Hughs P, Dodd C, Connell CR, Heins C, Kent SBH, Hood LE (1986) Fluorescent detection in automated DNA sequence analysis. Nature 321:673–681
Smith TJ, Blackman SA, Foster SJ (2000) Autolysins of Bacillus subtilis: multiple enzymes with multiple functions. Microbiology 146:249–262
Stewart AW, Johnson MG (1977) Increased numbers of heat-resistant spores produced by two strains of Clostridium perfringens bearing temperate phage s9. J Gen Microbiol 103:45–50
Sturino JM, Klaenhammer TR (2004) Bacteriophage defense systems and strategies for lactic acid bacteria. Adv Appl Microbiol 56:331–378
Van Immerseel F, De Buck J, Pasmans F, Huyghebaert G, Haesebrouck F, Ducatelle R (2004) Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol 33:537–549
Vieu JF, Guélin A, Dauguet C (1965) Morphology of the Welchia perfringens bacteriophage 80. Ann Inst Pasteur (Paris) 109:157–160
Webster R (1991) The tol gene products and the import of macromolecules into E. coli. Mol Microbiol 5:1005–1011
Wilgenbusch JC, Swofford D (2003) Inferring evolutionary trees with PAUP*. Curr Protoc Bioinform, Chapter 6: Unit 6.4
Wise MG, Siragusa GR (2005) Quantitative detection of Clostridium perfringens in the broiler fowl gastrointestinal tract by real-time PCR. Appl Environ Microbiol 71:3911–3916
Yan WK (1989) Use of host modified bacteriophages in development of a phage typing scheme for Clostridium perfringens. Med Lab Sci 46:186–193
Young R (2002) Bacteriophage holins: deadly diversity. J Mol Microbiol Biotechnol 4:21–36
Zimmer M, Scherer S, Loessner MJ (2002) Genomic analysis is Clostridium perfringes bacteriophage Φ3626, which integrates into guaA and possibly affects sporulation. J Bacteriol 184:4359–4368
Zimmer M, Vukov N, Scherer S, Loessner MJ (2002) The murein hydrolase of the bacteriophage Φ3626 dual lysis system is active against all tested Clostridium perfringes strains. Appl Environ Microbiol 68:5311–5317
Acknowledgments
The authors thank Mary Ard at the University of Georgia, College of Veterinary Medicine, for preliminary electron microscopy. The investigations were supported by the Agricultural Research Service, USDA CRIS project #6612-32000-046 (BSS, GRS, MS) and a non-funded CRADA #58-6612-7-175 with DEF at JCVI. The proteomics portion of the project was supported by NIH Grant Number P20 RR-016464 from the INBRE Program of the National Center for Research Resources (RW and KS). Sequencing was also supported by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (AK). The genome sequences were submitted to GenBank with accession numbers EU588980 for the ΦCP39O genome and GQ443085 for the ΦCP26F genome.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Seal, B.S., Fouts, D.E., Simmons, M. et al. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: genomic organization and proteomic analysis of the virions. Arch Virol 156, 25–35 (2011). https://doi.org/10.1007/s00705-010-0812-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-010-0812-z