Abstract
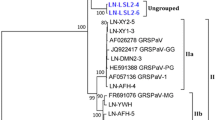
The complete genome sequences of two newly identified genetic variants of grapevine leafroll-associated virus 3 (GLRaV-3) are described. Two isolates, GH11 (18671 nt) and GH30 (18576 nt) were sequenced and found to be less than 70 % similar to other South African GLRaV-3 variants. The genome organization of GH11 and GH30 is similar to that of previously described GLRaV-3 isolates. However, no corresponding open reading frame 2 (ORF2) could be identified. Phylogenetic analysis indicates that GH11 and GH30 cluster in a subgroup (group VI) that is separate from the previously described GLRaV-3 isolates and should be regarded as a different strain of GLRaV-3.


Similar content being viewed by others
References
Pietersen G (2004) Spread of grapevine leafroll disease in South Africa—a difficult, but not insurmountable problem. Wynboer—a technical guide for wine producers. June 2004. http://www.wynboer.co.za/recentarticles/articles.php3. Accessed 17 Jan 2012
Martelli GP, Agranovsky AA, Bar-Joseph M, Boscia D, Candresse T, Coutts RHA, Dolja VV, Falk BW, Gonsalves D, Jelkmann W, Karasev AV, Minafra A, Namba S, Vetten HJ, Wisler GC, Yoshikawa N (2002) The family Closteroviridae revised. Arch Virol 147(10):2039–2044
Jooste AEC, Maree HJ, Bellstedt DU, Goszczynski DE, Pietersen G, Burger JT (2010) Three genetic grapevine leafroll-associated virus 3 variants identified from South African vineyards show high variability in their 5′UTR. Arch Virol 155:1997–2006
Maree HJ, Freeborough M-J, Burger JT (2008) Complete nucleotide sequence of a South African isolate of grapevine leafroll-associated virus 3 reveals a 5′UTR of 737 nucleotides. Arch Virol 153:755–757
Coetzee B, Freeborough M-J, Maree HJ, Celton J, Rees DJG, Burger JT (2010) Deep sequencing analysis of viruses infecting grapevines: virome of a vineyard. Virology 400(2):157–163
Zerbino DR, Birney E (2008) Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18(5):821–829
Osman F, Leutenegger C, Golino D, Rowhani A (2007) Real-time RT-PCR (TaqMan®) assays for the detection of grapevine leafroll associated viruses 1-5 and 9. J Virol Methods 141(1):22–29
White EJ, Venter M, Hiten NF, Burger JT (2008) Modified Cetyltrimethylammonium bromide method improves robustness and versatility: the benchmark for plant RNA extraction. Biotechnol J 3(11):1424–1428
Valverde RA, Nameth ST, Jordan RL (1990) Analysis of double-stranded-RNA for plant-virus diagnosis. Plant Dis 74(3):255–258
Meng B, Li C, Goszczynski DE, Gonsalves D (2005) Genome sequences and structures of two biologically distinct strains of Grapevine leafroll-associated virus 2 and sequence analysis. Virus Genes 31:31–41
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Gouveia P, Santos MT, Eiras-Dias JE, Nolasco G (2010) Five phylogenetic groups identified in the coat protein gene of grapevine leafroll-associated virus 3 obtained from Portuguese grapevine varieties. Arch Virol 156(3):413–420
Sharma AM, Wang J, Duffy S, Zhang S, Wong MK, Rashed A, Cooper ML, Daane KM, Almeida RPP (2011) Occurence of grapevine leafroll-associated virus complex in Napa valley. PLoS ONE 6(10):e26227. doi:10.1371/journal.pone.0026227
Ling KS, Zhu HY, Gonsalves D (2004) Complete nucleotide sequence and genome organization of grapevine leafroll-associated virus 3, type member of the genus Ampelovirus. J Gen Virol 85:2099–2102
Ling KS, Zhu HY, Drong RF, Slightom JL, Gonsalves D (1998) Nucleotide sequence of the 39-terminal two-thirds of the grapevine leafroll-associated virus-3 genome reveals a typical monopartite closterovirus. J Gen Virol 79:1299–1307
Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M (2008) CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res 37:205–210
Acknowledgments
The financial assistance of the National Research Foundation (NRF) towards this research is hereby acknowledged. Opinions expressed and conclusions arrived at are those of the authors and are not necessarily to be attributed to the NRF. The authors thank Winetech for research funding, Yolandi Nel for her contribution to the survey, and Dr. Dirk Stephan for critical reading of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bester, R., Maree, H.J. & Burger, J.T. Complete nucleotide sequence of a new strain of grapevine leafroll-associated virus 3 in South Africa. Arch Virol 157, 1815–1819 (2012). https://doi.org/10.1007/s00705-012-1333-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-012-1333-8