Abstract
We report a consistent set of AMBER force-field parameters for the most common phosphorylated amino acids, phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine in different protonation states. The calculation of atomic charges followed the original restrained electrostatic potential fitting procedure used to determine the charges for the parm94/99 parameter set, taking α-helical and β-strand conformations of the corresponding ACE-/NME-capped model peptide backbone into account. Missing force-field parameters were taken directly from the general AMBER force field (gaff) and the parm99 data set with minor modifications, or were newly generated based on ab initio calculations for model systems. Final parameters were validated by geometry optimizations and molecular-dynamics simulations. Template libraries for the phosphorylated amino acids in Leap format and corresponding frcmod parameter files are made available.

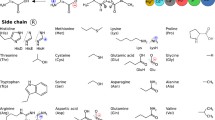
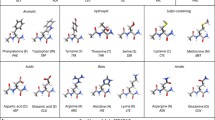
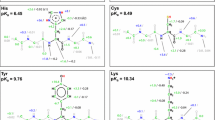
Schematic illustration of the systems used for parameter generation. Acid hydrogens are shown in red



Similar content being viewed by others
References
Cohen P (2002) Nat Cell Biol 4:E127–E130
Tholey A, Lindemann A, Kinzel V, Reed J (1999) Biophys J 76:76–87
Titgemeyer F, Hillen W (2002) Antonie Van Leeuwenhoek 82:59–71
Klumpp S, Krieglstein J (2002) Eur J Biochem 269:1067–1071
Rajagopal P, Waygood EB, Klevit RE (1994) Biochemistry 33:15271–15282
Cheatham TE III, Cieplak P, Kollman PA (1999) J Biomol Struct Dyn 16:845–862
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) J Am Chem Soc 117:5179–5197
Meagher KL, Redman LT, Carlson HA (2003) J Comput Chem 24:1016–1025
Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J (2001) Cell 105:115–126
Wozniak E, Oldziej S, Ciarkowski J (2000) Comp Chem 24:381–390
Bienkiewicz EA, Lumb KJ (1999) J Biomol NMR 15:203–206
Cieplak P, Cornell WD, Bayly C, Kollman PA (1995) J Comput Chem 16:1357–1377
Easton RE, Giesen DJ, Welch A, Cramer CJ, Truhlar DG (1996) Theor Chim Acta 93:281–301
Ditchfield R, Hehre WJ, Pople JA (1971) J Chem Phys 54:724–728
Frisch MJ, Pople JA, Binkley JS (1984) J Chem Phys 80:3265–3269
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2004) Gaussian 03. Gaussian Inc., Wallingford
Wang J, Cieplak P, Kollman PA (2000) J Comput Chem 21:1049–1074
Bayly CI, Cieplak P, Cornell WD, Kollman PA (1993) J Phys Chem 97:10269–10280
Case DA, Pearlman DA, Caldwell JW, Cheatham TE III, Wang J, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA (2002) Amber 7. University of California, San Francisco, CA
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Kosinsky YA, Volynsky PE, Lagant P, Vergoten G, Suzuki E, Arseniev AS, Efremov RG (2004) J Comput Chem 25:1313–1321
Hopfinger AJ, Pearlstein RA (1984) J Comput Chem 5:486–499
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) J Chem Phys 79:926–935
Bryce R, AMBER Parameter Database, http://pharmacy.man.ac.uk/amber/
Acknowledgements
The authors wish to thank Heike Meiselbach for helpful discussions, and the Leibnitz-Rechenzentrum in Munich and the Regionales Rechenzentrum Erlangen for computational resources. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB473, C10) to H. Sticht. N. Homeyer acknowledges a fellowship from the BioMedTec International Graduate School of Science (BIGSS), supported by the state of Bavaria.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Homeyer, N., Horn, A.H.C., Lanig, H. et al. AMBER force-field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model 12, 281–289 (2006). https://doi.org/10.1007/s00894-005-0028-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-005-0028-4