Abstract
The alginate lyase structural gene (AlyVI gene) was amplified from plasmid pET24-ALYVI carrying the alginate lyase gene from the marine bacterium Vibrio sp. QY101 which is a pathogen of Laminaria sp. When the gene was cloned into the multiple cloning site of the surface display vector pINA1317-YlCWP110 and expressed in cells of Yarrowia lipolytica, the cells displaying the alginate lyase could form clear zone on the plate containing sodium alginate, indicating that they had high alginate lyase activity. The cells displaying alginate lyase can be used to hydrolyze poly-β-d-mannuronate (M) and poly-α-l-guluronate (G) and sodium alginate to produce different lengths of oligosaccharides (more than pentasaccharides). This is the first report that the yeast cells displaying alginate lyase were used to produce different lengths of oligosaccharides from alginate.
Similar content being viewed by others
Introduction
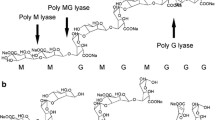
Alginate produced by brown seaweeds and certain bacteria is an anionic heteropolysaccharide which is composed of β-d-mannuronate (M) and α-l-guluronate (G) (Yoon et al. 2000). Because of the high viscosity and gelling properties of alginate with calcium ions, it is widely used in the food, pharmaceutical, and medical industries. The alginate hydrolysates and their derivatives exhibit many important bioactivities, such as stimulating human keratinocytes, accelerating plant root growth, increasing a growth rate of Bifidobacterium sp, producing protoplasts from brown sea algae, and enhancing penicillin production from cultures of Penicillium chrysogenum. Alginate oligosaccharides produced by alginate lyase can cause cytotoxic cytokine production in human mononuclear cells. The low-molecular-weight alginate derivatives also show antioxidation and prevention of cardiovascular and cerebrovascular diseases.
Alginate lyase splits the glycoside linkages of alginate chains via β-elimination to produce oligosaccharides (Yoon et al. 2000). Many marine bacteria, such as Pseudoalteromonas elyakovii IAM (Sawabe et al. 2001), Sphingomonas sp. A1 (Miyake et al. 2003), Klebsiella pneumoniae (Caswell et al. 1989), Vibrio sp. QY101 (Han et al. 2004), Vibrio sp. YKW-34 (Fu et al. 2007), and Pseudomonas sp. OS-ALG-9 (Fujiyama et al. 1995), have been found to produce a large amount of alginate lyases. So far, approximately 20 genes encoding alginate lyases from marine bacteria have been cloned and sequenced. A1-IV from Sphingomonas sp. A1 (Miyake et al. 2003), alginate lyases from K. pneumoniae (Caswell et al. 1989), Pseudomonas sp. OS-ALG-9 (Fujiyama et al. 1995), Vibrio sp. QY101 (Han et al. 2004), Corynebacterium strain ALY-1 (Matsubara et al. 2000), Deleya marina (Kraiwattanapong et al. 1999), P. elyakovii IAM (Sawabe et al. 2001; Ma et al. 2008), and Streptomyces sp. ALG-5 (Kim et al. 2008) were successfully expressed in Escherichia coli cells. It has been reported that the marine bacterium Vibrio sp. QY101 which was isolated from a decaying thallus of Laminaria sp. is a pathogen of it (Han et al. 2004). Therefore, it is not feasible to produce alginate lyase on large scale using the pathogenic bacterium.
In recent years, yeast surface display techniques have received increasing attention as they have many applications in biotechnological and industrial fields, such as cell adhesion, molecular recognition, immobilized biocatalysis, bioconversion, bioremediation, change of cell function, signal transduction, biosensor, live vaccine development and ultra-high-throughput screening for the identification of novel biocatalysts (Yue et al. 2008; Zhu et al. 2006; Ni et al. 2008). Alkaline protease, inulinase, amylases, cellulases, xylanases, hemolysin, and other proteins have been successfully immobilized on yeast cells and their potential applications have been evaluated (Yue et al. 2008; Zhu et al. 2006; Ueda and Tanaka 2000; Ni et al. 2008). However, alginate lyase has not been displayed on yeast cells for hydrolysis of alginate so far. A surface display vector pINA1317-YlCWP110 for protein display on the yeast Yarrowia lipolytica was constructed in our laboratory (Yue et al. 2008). The vector has many advantages over other surface display vectors. It uses a strong recombinant growth-phase-dependent promoter, hp4d (Madzak et al. 2004). The vector carries zeta elements, which allow them to integrate genomic DNA in Y. lipolytica strains which are generally regarded as safe. Furthermore, only a “yeast expression cassette” is used for the transformation of the recipient strain. The resulting strain is devoid of bacterial DNA, retaining generally regarded as safe status and avoiding the spread of antibiotic resistance genes in the environment.
Because the alginate-lyase-producing marine bacteria reported so far are the pathogen to Laminaria sp, it is not feasible to use it for production of the enzyme by the bacteria. The enzyme displayed on the cells of Y. lipolytica can be reused and regenerated and is “naturally” immobilized on the cell surface. Therefore, in this study, the alginate lyase gene from the marine bacterium Vibrio sp. QY101 was cloned into pINA1317-YlCWP110 and expressed in cells of Y. lipolytica. The expressed alginate lyase was displayed on the yeast cells. The yeast cells displaying the alginate lyase were used to hydrolyze alginate.
Materials and Methods
Strains and media
The yeast Y. lipolytica Po1h (MatA, ura3-302, xpr2-322, axp1-2; phenotype: Ura−, AEP, AXP, Suc+; Madzak et al. 2004) was used as a host for cell surface display. The yeast strain was grown in yeast–polypeptone–dextrose; 1.0% yeast extract, 2.0% bacto-peptone, 2.0% glucose). The yeast transformants were selected on YNB-N5000 (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 1.0% glucose, 0.5% ammonium sulfate). In order to produce displayed protein by yeast transformants, the PPB medium was used (Jolivalt et al. 2005). The E. coli strain used in this study for plasmid recovery and cloning experiments was DH5α [F− endA1 hsdR17(rK−/mK+)supE44 thi-1λ− recA1 gyr96ΔlacU169(ϕ80lacZΔM15)] and was grown in Luria broth (LB). The E. coli transformants were grown in LB medium with 30 μg/ml of kanamycin.
Plasmids
pET24-ALYVI (Kanr; Han et al. 2004) carrying the alginate lyase gene from the marine bacterium Vibrio sp. QY101 which was isolated from a decaying thallus of Laminaria sp. was used as the template for amplification of the alginate lyase gene by polymerase chain reaction (PCR). The surface display vector pINA1317-YlCWP110 which contains the C-terminal end of YlCWP1 from Y. lipolytica was constructed in this laboratory (Yue et al. 2008). pMD19-T was purchased from TaKaRa (Japan).
Isolation of DNA, restriction digestions, and transformation
DNA manipulations were carried out using standard methods (Sambrook et al. 1989). Bacterial plasmid DNA was purified using Perfectprep plasmid minikit (Eppendorf). Restriction endonuclease digestions and DNA ligations were performed according to the manufacturer’s recommendations. E. coli was transformed with plasmid DNA according to Sambrook et al. (1989). Transformants were plated out onto LB medium containing 30 μg/ml of kanamycin. Y. lipolytica was transformed according to the methods as described by Xuan et al. (1988).
DNA sequence and computer analysis
BLAST and ORF Finder programs at the National Center for Biotechnology Information were used for the nucleotide sequence analysis, deduction of the amino acid sequence, and database searches.
Amplification of the alginate lyase gene from pET24-ALYVI (Kanr)
The primers for amplification of the gene encoding the alginate lyase were designed according to ALYVI gene (accession number: AY221030) and the multiple cloning site of the surface display vector pINA1317-YlCWP110 (Yue et al. 2008). The forward primer Aly1 was 3′-GGCCGTTCTGGCCGAACAAGCAGAGCTTAAAAC-5′ (the bases underlined encode SfiI restriction site) and the reverse primers (Aly2 and Aly3) were 3′-CGGCGCGGATCCTCTATGATCAATATTGATCTTCG-5′ (the bases underlined encode BamHI restriction site) and 
 (the bases underlined encode BamHI restriction site and the bases boxed encode 6-His), respectively. The gene encoding the alginate lyase was amplified by PCR using pET24-ALYVI as a template. The reaction system (50 μl) was composed of 5.0 μl of TaKaRa 10× Ex Taq buffer, 4.0 μl of TaKaRa 5 mM dNTP, 2.0 μl (20 μM) of Aly1, 2.0 μl (20 μM) of Aly2 or Aly3, 0.3 μl of ExTaq DNA polymerase (5 U/μl), 2.0 μl (10.0 ng/ml) of the template DNA, and 35.0 μl of H2O. The conditions for the PCR amplification were as follows: initial denaturation at 94°C for 5 min, denaturation at 94°C for 45 s, annealing temperature at 62°C for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 8 min. PCR was run for 30 cycles and the PCR cycler was GeneAmp PCR System 2400 (PerkinElmer, Waltham, MA, USA). PCR products were separated in agarose gel and recovered using TaKaRa Agarose Gel DNA Fragment Recovery Kit Ver.2.0. The recovered PCR products were ligated into pMD19-T and transformed into the competent cells of E. coli DH5α. The transformants were selected on the plates with kanamycin. The plasmids were extracted by using the methods as described by Sambrook et al. (1989).The cloned DNA fragments inserted on the vector were sequenced by Shanghai Sangon Company. After alignment, it was found that the sequence of the cloned DNA fragments was completely identical to the DNA sequence of the ALYVI gene (accession number: AY221030).
(the bases underlined encode BamHI restriction site and the bases boxed encode 6-His), respectively. The gene encoding the alginate lyase was amplified by PCR using pET24-ALYVI as a template. The reaction system (50 μl) was composed of 5.0 μl of TaKaRa 10× Ex Taq buffer, 4.0 μl of TaKaRa 5 mM dNTP, 2.0 μl (20 μM) of Aly1, 2.0 μl (20 μM) of Aly2 or Aly3, 0.3 μl of ExTaq DNA polymerase (5 U/μl), 2.0 μl (10.0 ng/ml) of the template DNA, and 35.0 μl of H2O. The conditions for the PCR amplification were as follows: initial denaturation at 94°C for 5 min, denaturation at 94°C for 45 s, annealing temperature at 62°C for 45 s, extension at 72°C for 1 min, and final extension at 72°C for 8 min. PCR was run for 30 cycles and the PCR cycler was GeneAmp PCR System 2400 (PerkinElmer, Waltham, MA, USA). PCR products were separated in agarose gel and recovered using TaKaRa Agarose Gel DNA Fragment Recovery Kit Ver.2.0. The recovered PCR products were ligated into pMD19-T and transformed into the competent cells of E. coli DH5α. The transformants were selected on the plates with kanamycin. The plasmids were extracted by using the methods as described by Sambrook et al. (1989).The cloned DNA fragments inserted on the vector were sequenced by Shanghai Sangon Company. After alignment, it was found that the sequence of the cloned DNA fragments was completely identical to the DNA sequence of the ALYVI gene (accession number: AY221030).
Alginate lyase display on cells of Y. lipolytica
The gene amplification by PCR, the gene expression, and the lyase display on the yeast cells of Y. lipolytica were performed as described by Yue et al. (2008). The pMD19-T carrying the cloned ALYVI gene was digested with SfiI at 50°C overnight then with BamHI at 37°C for 3 h. The digests containing the ALYVI gene were separated in agarose gel and recovered using TaKaRa Agarose Gel DNA Fragment Recovery Kit Ver.2.0. The recovered digest products were ligated into the surface display vector pINA1317-YlCWP110 digested with the same enzymes (Yue et al. 2008) and the resulting plasmid carrying ALYVI gene was designated as pINA1317-YlCWP110-ALY (Fig. 1). The plasmid was transformed into the competent cells of E. coli DH5α. The transformants were selected on the plates with kanamycin. The plasmids were extracted by using the methods as described by Sambrook et al. (1989). pINA1317-YlCWP110-ALY extracted from E. coli transformants were digested with NotI. The fragments carrying YlCWP110-ALY were separated in agarose gel and recovered using TaKaRa Agarose Gel DNA Purification Kit Ver.2.0. The recovered fragments were transformed into Y. lipolytica Po1h by lithium acetate method (Xuan et al. 1988). The transformants were spread on the YNB plates and grown at 28°C for 96 h. Y. lipolytica Po1h only carrying fragment YlCWP110 was used as a control.
Integration of the ALYVI gene into genomic DNA in the positive transformants was confirmed as described below. The positive transformants carrying ALYVI gene were grown in PPB liquid medium for 120 h. The alginate lyase activity of different positive transformants was determined as described below. The cells of Y. lipolytica Po1h only carrying yeast cassette without ALYVI gene were used as a control.
Determination of alginate lyase activity
The cells of the positive transformants carrying ALYVI gene and Y. lipolytica Po1h only carrying yeast cassette without ALYVI gene were grown in PPB medium for 120 h, respectively. The cultures were washed three times with phosphate-buffered saline (PBS) buffer (pH 7.2) by centrifugation at 6,000×g and 4°C for 5 min. The pellets were resuspended in PBS buffer (pH 7.2). Half of milliliter of the cell suspension was mixed with 0.5 ml of 0.3% sodium alginate solution (dissolved in PBS buffer pH 7.2) and the mixture was incubated at 30°C for 30 min. After that, the reaction was stopped immediately in boiling water for 5 min. After cooling, the mixture was centrifuged at 14,000×g for 10 min and optical density value of the supernatant obtained was determined with spectrophotometer at 235 nm (Li et al. 2003). The reaction mixtures which contained the same cell suspension inactivated at 100°C for 5 min were used as a control. One unit of alginate lyase activity was defined as the increase of 0.01 absorbance unit at 235 nm. Cell dry weight of the cultures was determined as described by Gao et al. (2007). The specific alginate lyase activity was defined as units per gram of cell dry weight. Alginate lyase activity was also estimated using the plate which contained 0.5% sodium alginate. Different colonies of the transformants were transferred to the plates and incubated at 28°C for 5 days. Then, 70% ethanol solution was added to the plates and the colonies were submerged for 30 min. Finally, the clear zones around the colonies were observed and photographed.
Effects of pH and temperature on the displayed alginate lyase activity
The effect of pH on the alginate lyase activity was determined by incubating the yeast cells displaying alginate lyase between pH 4.0 and 9.0 using the standard assay condition. The buffers used were 0.1 M acetate buffer (pH 4.0–6.0) and 0.1 M Tris–HCl buffer (pH 7.0–9.0). The optimal temperature for activity of the alginate lyase was determined at temperatures of 16°C, 30°C, 40°C, and 50°C in 0.1 M PBS buffer (pH 7.3).
Immunofluorescence microscopy
The immunofluorescence microscopy was conducted as described by Adams et al. (1998) using the 6× His monoclonal antibody produced by Clontech (USA) as primary antibody, and immunoglobulin G/fluorescein isothiocyanate (IgG/FITC) as secondary antibody produced by ZSGB-BIO (China). Y. lipolytica Po1h cells carrying YlCWP110-ALY were grown in PPB liquid medium for 120 h. The cells were collected and washed three times by centrifugation with PBS buffer. The pellets were suspended in 3.7% formaldehyde and incubated overnight at 28°C with shaking. Then, the yeast cells were labeled using the primary and secondary antibodies mentioned above.
Confirmation of integration of the target gene into Y. lipolytica genome
To get the evidence that DNA fragments carrying YlCWP110-ALY have been integrated into Y. lipolytica genome, genomic DNAs from the corresponding transformants were extracted and used as templates for PCR. The forward primer and the reverse primer used for this checking were Aly1 and Aly2 as described above, respectively. PCR amplification was performed as described above. The sizes of the PCR products were estimated using the Automated Gel Documentation and Analysis System (Gene-Genius, USA).
Depolymerization of sodium alginate, polyM, and polyG by the displayed alginate lyase
The mixture consisting of 0.3% (w/v) sodium alginate or 0.3% (w/v) polyG or 0.3% (w/v) polyM, 0.1 M PBS buffer (pH 7.3), and 200 μl of the yeast cells displaying the alginate lyase was incubated at 30°C for 4 h. The alginate hydrolysates released were determined as described above.
Thin-layer chromatography
The alginate-depolymerization products were analyzed by thin-layer chromatography (TLC) using a solvent system of 1-butanol–acetate acid–water (3:2:3, v/v). The products were visualized by heating the TLC plate at 130°C for 2 min after spraying with 10% (v/v) sulfuric acid in ethanol (Hashimoto et al. 2000).
Results and discussion
Effects of 6×His Tag on the activity of alginate lyase immobilized on the yeast cells
The ALYV I gene encoding alginate lyase from the marine bacterium Vibrio sp. QY101 was amplified from the plasmid pET24-ALYVI (Kanr) carrying the alginate lyase gene with the reverse primers with the bases encoding 6× His tag and without the bases encoding 6× His tag. Construction of the plasmid pINA1317-YlCWP110-ALY for the alginate lyase surface display on Y. lipolytica cells is shown in Fig. 1. After the genes were cloned into the surface display plasmid pINA1317-YlCWP110, they were expressed in the yeast cells of Y. lipolytica and the alginate lyase was displayed on the yeast cells. We found that the activity of the alginate lyase with 6× His tag was much higher than that of the alginate lyase without 6× His tag. For example, the transformant His7 displaying the alginate lyase with 6× His tag had the specific activity of over 208.0 U/g of cell dry weight (Fig. 2a) while the transformant 3 displaying the alginate lyase without 6× His tag had the specific activity of less than 85.4 U/g of cell dry weight (Fig. 2b). The colonies carrying the alginate lyase with 6× His tag also could form bigger clear zones on the plates than those carrying the alginate lyase without 6× His tag (Fig. 3). Thus, the transformant His7 displaying the alginate lyase with 6× His tag was used for the subsequent studies. Strauss and Gotz (1996) also reported that the activity of surface-immobilized lipase varied with the spacer length. The activity increases from 0.8 to 83 units per milligram lipase as the spacer length varies from 10 to 92 amino acids. They thought that the length of the cell-wall-spanning region of the carrier protein must exceed a critical length to allow efficient folding of the passenger protein. Therefore, 6× His tag as the spacer of the displayed alginate lyase on the yeast cells may also play positive role in the efficient folding of the alginate lyase on the yeast cells, resulting in the increased alginate lyase activity (Figs. 2 and 3). However, it is still unknown if the longer His tag could further increase alginate lyase activity.
The clear zones (A–D) formed on the plates with sodium alginate by the transformants carrying ALYV I gene and no clear zone (E) formed by the control cells only carrying YlCWP-110 without ALYV I gene. The clear zones (A and D) were formed by the transformants carrying ALYV I gene without the bases encoding 6× His tag while the clear zones (B and C) were formed by the transformants carrying ALYV I gene with the bases encoding 6× His tag
In order to confirm the presence of the 6× His-alginate lyase-YlCWP110 fusion protein on the yeast cell surface, immunofluorescence labeling of the yeast cells was performed with 6× His monoclonal antibody as primary antibody and IgG/FITC as secondary antibody. YlCWP110-ALY-carrying Y. lipolytica cells were grown in PPB medium for 120 h. The Y. lipolytica cells displaying 6× His-alginate lyase-YlCWP110 fusion protein were labeled by IgG/FITC, and green fluorescence on the yeast cells was observed, while no fluorescence was observed on control YlCWP110-carrying Y. lipolytica cells (Fig. 4). These results clearly demonstrated that the 6× His-alginate lyase-YlCWP110 fusion protein was displayed on the cell surface, allowing its recognition by the antibodies. It can also be noticed from the results in Fig. 4 that 100% of the observed cells displayed alginate lyase. In our previous studies (Yue et al. 2008; Ni et al. 2008), it was also found that 100% of the Y. lipolytica cells displayed enhanced green fluorescent protein, hemolysin, and alkaline protease using the same glycosylphosphatidyl inositol-anchor-fusion expression system and the Y. lipolytica cells displaying hemolysin and alkaline protease exhibited hemolytic activity toward erythrocytes from flounder and all the proteins tested, respectively.
Immunofluorescent labeling of transformed Y. lipolytica cells using 6× His monoclonal antibody as primary antibody and IgG/FITC as secondary antibody. Microphotographs were taken under visible light (b and d), and immunofluorescence microphotographs were taken under emission at 550 nm (a and c). c and d: Y. lipolytica control cells harboring YlCWP110; a and b: Y. lipolytica cells harboring YlCWP110-ALY. Magnification 40 × 10
After the fragments without the signal sequence of ALYVI gene were amplified from genomic DNA in Y. lipolytica carrying the alginate lyase gene by PCR, the expected size of PCR product of 1,210 bp in size was observed (data not shown), suggesting that ALYVI gene indeed has been integrated into the genomic DNAs in Y. lipolytica.
Optimal pH and temperature of the alginate lyase displayed on the yeast cells
The alginate lyase activity was measured at various pHs in buffers with the same ionic concentrations. The maximum activity (363 U/g) of the alginate lyase displayed on the yeast cells was observed at pH 7.3 (Fig. 5). However, the optimal pH of the purified recombinant alginate lyase from E. coli BL21 (DE3) cells containing the plasmid carrying the alginate lyase gene and the purified native alginate lyase produced by Vibrio sp. QY101 was found to be 7.5, respectively (Han et al. 2004; Li et al. 2003). The maximum activity of the displayed alginate lyase, 360 U/g, was observed at 40°C during the tested temperature ranging from 15°C to 50°C (Fig. 6). The optimal temperature of the purified recombinant alginate lyase from E. coli BL21 (DE3) cells containing plasmid was also 40°C (Han et al. 2004). This indicates that the optimal temperature and pH of the alginate lyase immobilized on the yeast cells were the same as those of the recombinant alginate lyase produced by E. coli cells carrying pET24-alyVI. In contrast, the optimal temperature of the purified alginate lyase from Vibrio sp. QY101 was 30°C (Li et al. 2003). However, the optimal temperature of the alkaline protease was decreased after it was immobilized on the yeast cells by the same way compared to that of the free alkaline protease purified from Aureobasidium pullulans HN2.3 (Ni et al. 2008).
Hydrolysis of alginate, polyM, and polyG by the displayed alginate lyase
Based on the substrate specificity, alginate lyases have been classified as poly(M) lyases (EC 4.2.2.3) and poly(G) lyases (EC 4.2.2.11), which preferentially degrade the poly(M) block and poly(G) block, respectively. Therefore, sodium alginate, polyM, and polyG were digested by the displayed alginate lyase and the alginate hydrolysates released from sodium alginate, polyM, and polyG were determined as described in “Materials and methods,” respectively. The results in Fig. 7 show that polyM can be much more easily hydrolyzed by the displayed alginate lyase than polyG and sodium alginate. This means that the displayed alginate lyase is poly(M) lyases. Alginate lyases from herbivorous marine mollusks such as abalone and turban shell are also poly(M) lyases (Yamamoto et al. 2008). A1-IV’and A1-IV from Sphingomonas sp. prefer polyM (Hashimoto et al. 2005). However, recently, bifunctional alginate lyases which act on both poly(M) and poly(G) blocks have also been isolated. For example, the extracellular alginate lyase produced by P. elyakovii and the recombinant E. coli is capable of degrading all block structures derived from sodium alginate and produces a series of tri to octaoligouronates (Sawabe et al. 2001; Ma et al. 2008). In contrast, A1-II from Sphingomonas sp. prefers polyG rather than polyM and releases tri and tetrasaccharides (Yoon et al. 2000). The alginate lyase produced by Streptomyces sp. ALG-5 appears to be poly-guluronate lyase degrading polyG block preferentially than polyM block (Kim et al. 2008).
The hydrolysis products of alginate, polyM, and polyG by the displayed alginate lyase were analyzed by TLC. All the alginate, polyM, and polyG could be converted into different lengths of oligosaccharides (more than pentasaccharides) by the displayed alginate lyase (Fig. 8). This characteristic may find highly potential applications in seaweed genetics, food, and pharmaceutical industries. Alginate that was purified from the fermentation solution of marine bacterium Vibrio sp. 510 under specific reaction conditions was hydrolyzed by the native alginate lyase and seven oligosaccharides, including di, tri, and tetrasaccharides, were detected (Zhang et al. 2004). All the alginate, polyM, and polyG can be converted into oligosaccharides with more than tetrasaccharides by the purified recombinant alginate lyase from E. coli (Ma et al. 2008). The degraded products by alginate lyase produced by Streptomyces sp. ALG-5 were found to be di, tri, tetra, and pentasaccharides (Kim et al. 2008).
Thin-layer chromatogram of the end products of alginate, polyM, and polyG after hydrolysis with the displayed alginate lyase. Lane 1 glucose; lane 2: maltose; lane 3: raffinose; lane 4: unhydrolyzed polyM (1.0%); lane 5: unhydrolyzed polyG (1.0%); lane 6: unhydrolyzed alginate (1.0%); lane 7: hydrolyzed polyM (1.0%); lane 8: hydrolyzed polyG (1.0%) and 7: hydrolyzed alginate (1.0%). The end products of the hydrolysis were analyzed by using TLC plate as described in “Materials and Methods.” The products were visualized by heating the TLC plate after spraying with 10% (v/v) sulfuric acid in ethanol
References
Adams A, Gottschling DE, Kaiser CA, Stearms T (1998) Yeast immunofluorescence. In: Burke D, Dawson D (eds) Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory, New York, p 100
Caswell RC, Gacesa P, Lutrell KF, Weightam AJ (1989) Molecular cloning and heterologous expression of a Klebsiella pneumoniae gene encoding alginate lyase. Gene 75:127–134
Fu XT, Lin H, Kim SM (2007) Purification and characterization of a Na+/K+ dependent alginate lyase from turban shell gut Vibrio sp. YKW-34. Enzy Microb Technol 41:828–834
Fujiyama K, Maki H, Kinoshita S, Yoshida T (1995) Purification and characterization of the recombinant alginate lyase from Pseudomonas sp. leaked by Escherichia coli upon addition of glycine. FEMS Microbiol Lett 126:19–25
Gao LM, Chi ZM, Sheng J, Ni XM, Wang L (2007) Single-cell protein production from Jerusalem artichoke extract by a recently isolated marine yeast Cryptococcus aureus G7a and its nutritive analysis. Appl Microbiol Biotechnol 77:825–832
Han F, Gong QH, Song K, Li JB, Yu WG (2004) Cloning, sequence analysis and expression of gene AlyVI encoding alginate lyase from marine bacterium Vibrio sp. QY101. DNA Sequence 0:1–8
Hashimoto W, Miyake O, Momma K, Kawai S, Murata K (2000) Molecular identification of oligoalginate lyase of Sphingomonas sp. strain A1 one of the enzymes required for complete depolymerization of alginate. J Bacteriol 182:4572–4577
Hashimoto W, Miyake O, Ochiai A, Murata K (2005) Molecular identification of Sphingomonas sp. A1 alginate lyase (A1-IV) as a member of novel polysaccharide lyase family 15 and implications in alginate lyase evolution. J Biosci Bioeng 99:48–54
Jolivalt C, Madzak C, Brault A, Caminade E, Malosse C, Mougin C (2005) Expression of laccase lllb from the white-rot fungus Trametes versicolor in the yeast Yarrowia lipolytica for environmental applications. Appl Microbiol Biotechnol 66:450–456
Kim DE, Lee EY, Kim HS (2008) Cloning and characterization of alginate lyase from a marine bacterium Streptomyces sp. ALG-5. Mar Biotechnol. doi:10.1007/s10126-008-9114-9
Kraiwattanapong J, Tsuruga H, Ooi T, Kinoshita S (1999) Cloning and sequencing of a Deleya marina gene encoding for alginate lyase. Biotechnol Lett 21:169–174
Li JB, Yu WG, Han F, Han WJ, Song K (2003) Purification and characterization of alginate lyase from the marine bacterium Vibrio sp. QY101. J Microbiol Sin 4:753–757
Ma LY, Chi ZM, Li J, Wu LF (2008) Overexpression of alginate lyase of Pseudoalteromonas elyakovii in Escherichia coli, purification, and characterization of the recombinant alginate lyase. World J Microbiol Biotechnol 24:89–96
Madzak C, Gaillardin C, Beckerich JM (2004) Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol 109:63–81
Matsubara Y, Kawada R, Iwasaki K, Kimura Y, Oda T, Muramatsu T (2000) Cloning and sequence analysis of a gene (aly PG) encoding poly(a-L-guluronate) lyase from Corynebacterium sp. strain ALY-1. J Biosci Bioeng 89:199–202
Miyake O, Hashimoto W, Murata K (2003) An exotype alginate lyase in Sphingomonas sp. A1: overexpression in Escherichia coli, purification, and characterization of alginate lyase IV (A1-IV). Protein Expr Purif 29:33–41
Ni XM, Yue LX, Chi ZM, Li J, Wang XH, Madzak C (2008) Alkaline protease gene cloning from the marine yeast Aureobasidium pullulans HN2-3 and the protease surface display on Yarrowia lipolytica for bioactive peptide production. Mar Biotechnol. doi:10.1007/s10126-008-9122-9
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York, p 367 – 370 (Chinese translated edn.)
Sawabe T, Takahashi H, Ezura Y, Gacesa P (2001) Cloning, sequence analysis and expression of Pseudoalteromonas elyakovii IAM 14594 gene (alyPEEC) encoding the extracellular alginate lyase. Carbohydr Res 335:11–21
Strauss A, Gotz F (1996) In vivo immobilization of enzymatically active polypeptides on the cell surface of Staphylococcus carnosus. Mol Microbiol 21:491–500
Ueda M, Tanaka A (2000) Genetic immobilization of proteins on the yeast cell surface. Biotechnol Adv 18:121–140
Xuan JM, Fournier P, Gaillardin C (1988) Cloning of the LYS5 gene encoding saccharopine dehydrogenase. Curr Genet 14:15–21
Yamamoto S, Sahara T, Sato D, Kawasaki K, Ohgiya S, Inoue A, Ojima T (2008) Catalytically important amino-acid residues of abalone alginate lyase HdAly assessed by site-directed mutagenesis. Enzy Microb Technol 43:396–402. doi:10.1016/j.enzmictec.2008.06.006
Yoon HJ, Hashimoto W, Miyake O, Okamoto M, Mikami B, Murata K (2000) Overexpression in Escherichia coli, purification, and characterization of Sphingomonas sp. A1 alginate lyases. Protein Expr Purif 19:84–90
Yue LX, Chi ZM, Wang L, Liu J, Madzak C, Li J, Wang XH (2008) Construction of a new plasmid for surface display on cells of Yarrowia lipolytica. J Microbiol Meth 72:116–123
Zhang Z, Yu G, Guan H, Zhao X, Du Y, Jiang X (2004) Preparation and structure elucidation of alginate oligosaccharides degraded by alginate lyase from Vibrio sp. 510. Carbohydr Res 339:1475–1481
Zhu KL, Chi ZM, Li J, Zhang FL, Li MJ, Yasoda HN, Wu LF (2006) The surface display of haemolysin from Vibrio harveyi on yeast cells and their potential applications as live vaccine in marine fish. Vaccine 24:6046–6052
Acknowledgements
This research was supported by grant 30771645 from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, G., Yue, L., Chi, Z. et al. The Surface Display of the Alginate Lyase on the Cells of Yarrowia lipolytica for Hydrolysis of Alginate. Mar Biotechnol 11, 619–626 (2009). https://doi.org/10.1007/s10126-009-9178-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-009-9178-1