Abstract
The purpose of our study is to identify and quantify the association between high breast mass density and breast malignancy using inductive logic programming (ILP) and conditional probabilities, and validate this association in an independent dataset. We ran our ILP algorithm on 62,219 mammographic abnormalities. We set the Aleph ILP system to generate 10,000 rules per malignant finding with a recall >5% and precision >25%. Aleph reported the best rule for each malignant finding. A total of 80 unique rules were learned. A radiologist reviewed all rules and identified potentially interesting rules. High breast mass density appeared in 24% of the learned rules. We confirmed each interesting rule by calculating the probability of malignancy given each mammographic descriptor. High mass density was the fifth highest ranked predictor. To validate the association between mass density and malignancy in an independent dataset, we collected data from 180 consecutive breast biopsies performed between 2005 and 2007. We created a logistic model with benign or malignant outcome as the dependent variable while controlling for potentially confounding factors. We calculated odds ratios based on dichomotized variables. In our logistic regression model, the independent predictors high breast mass density (OR 6.6, CI 2.5–17.6), irregular mass shape (OR 10.0, CI 3.4–29.5), spiculated mass margin (OR 20.4, CI 1.9–222.8), and subject age (β = 0.09, p < 0.0001) significantly predicted malignancy. Both ILP and conditional probabilities show that high breast mass density is an important adjunct predictor of malignancy, and this association is confirmed in an independent data set of prospectively collected mammographic findings.
Similar content being viewed by others
Introduction
The increasingly computerized nature of modern medicine creates vast amounts of biomedical data as a result of patient care. Large amounts of data are generated in radiology not only due to data-rich image files but because of data available in structured imaging reports, such as those utilized in breast imaging. In the past, these repositories of data have been used to generate novel hypotheses using data mining techniques for knowledge discovery.1–3 After hypotheses have been generated, however, an integral part of the knowledge discovery process is evaluation of the discovered knowledge and its application in healthcare.4 One important aspect of this evaluation is assessing the generalizability of the results outside of the dataset that generated the knowledge. Validating the findings of inductive logic programming (ILP) within the domain of breast imaging is the focus of this project.
ILP5 is a computer programming technique that is particularly helpful in aiding researchers in the data mining and knowledge discovery process. ILP's objective is to discover a set of if–then rules that predict the presence or absence of a disease state or outcome. Generating ILP rules requires: (1) a series of positive and negative examples, (2) background knowledge about those examples, and (3) user-defined constraints such as performance parameters about what type of rules may be learned. ILP has several advantages over propositional learning techniques such as logistic regression. First, it can utilize data from a relational database with many tables, rather than a single table. Second, the discovered rules, which are based in logic, are easily understood by both humans and computers. Finally, the rules can provide insight about predictive indicators that distinguish the negative examples from the positive examples.
One method to confirm the association between a predictive indicator and a specific outcome is probability theory.6 Conditional probabilities describe the probability of a specific outcome given a set of indicators, for example the probability of malignancy given a finding on a mammogram. Such probabilities provide insight into indicators that are more or less predictive of a specific outcome. By using ILP-generated rules in conjunction with probability theory, we can identify and confirm novel indicators for later targeted evaluation and application.
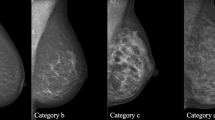
Breast imaging is a subspecialty area of radiology ideally suited to hypothesis generation using data mining techniques. First, there is a standardized lexicon, the Breast Imaging Reporting and Data System (BI-RADS),7 which is used by radiologists when interpreting mammograms. BI-RADS encourages radiologists to use specified language to describe mammographic findings (Fig. 1), which in turn can be used to generate evidence-based recommendations. Research has shown that BI-RADS descriptive terms can make accurate and reliable predictions of breast cancer.8–16 Second, many breast radiologists report their findings on a structured report, using reporting software. The databases behind these structured reports can be queried for the mammographic findings, and the data can be mined to generate novel hypotheses.
The predictive capability of one BI-RADS descriptor, the mammographic density of a mass, remains controversial. Mammographic mass density is determined by comparing the density of the mass to an equal volume of breast fibroglandular tissue. The BI-RADS categories for mass density are fat-containing, low density, equal (“iso”) density, and high density (Fig. 1). For example, if the mass is higher density (more white appearing on the mammogram), it is recorded as “high density.” In the past, experts have asserted, with limited scientific data, that high-density masses are more likely malignant. The only study reported in the literature to evaluate the association between breast mass density and cancer explicitly showed that mass density is difficult to consistently evaluate and that breast mass density contributes less to predicting malignancy than traditionally thought.17 Since that research was performed in the early 1990s, no studies in the literature have evaluated the contribution of breast mass density to the prediction of cancer. Recently, however, research using ILP showed that high mass density may indeed be an important predictor of cancer.1 The purpose of this study is to confirm the conclusions of this previous research using a modified ILP method and probability theory, and then to validate this conclusion by specifically assessing the association between breast mass density and pathologic outcome in an independent dataset of mammographic findings.
Materials and Methods
The Institutional Review Board of the University of Wisconsin School of Medicine and Public Health approved and waived informed consent for this study. The study fully complied with the Health Insurance Portability and Accountability Act.
Knowledge Discovery
This study involved using a modified ILP algorithm in conjunction with probability theory, on a series of mammographic abnormalities linked to the Wisconsin Cancer Reporting System, the state's breast cancer registry. This exploratory dataset has been described previously in the literature,1 but it is briefly described below for the convenience of the reader.
Dataset of Positive and Negative Examples
Our series of positive and negative examples, the exploratory dataset, utilized by the ILP algorithm contains 48,744 mammograms from 18,270 patients performed at the Froedtert and Medical College of Wisconsin Breast Imagining Center between April 5, 1999, and February 9, 2004. All of the 62,219 mammographic abnormalities were matched with the state cancer registry. The dataset contains a total of 510 malignant and 61,709 benign abnormalities.
Background Knowledge
Background knowledge consists of all information about the 62,219 abnormalities. The background knowledge for our exploratory dataset contains descriptive indicators about the findings on the mammogram, patient risk factors such as age and family history, and the radiologist's BI-RADS final assessment of each abnormality as shown in Table 1. Each abnormality was interpreted by one of eight radiologists, two of which were fellowship-trained, and the background knowledge was collected by querying a mammography structured reporting and tracking data system (PenRad Technologies®, Minnetonka, MN, USA). In addition, the background knowledge for our analysis also included variables for other findings on the same mammogram, and other findings on the patient's previous mammograms.
User-defined Constraints
User-defined constraints limit the number and type of rules that are generated by the ILP algorithm. We limited the number of rules considered to 10,000 per malignant finding, and retained rules that had a recall of at least 5% and a precision of at least 25%. For the purposes of our study, recall is defined as the fraction of truly malignant abnormalities that a rule labels as malignant (analogous to sensitivity). Precision is defined as the fraction comprised of malignant abnormalities that are accurately labeled as malignant by a rule (analogous to positive predictive value).
Each rule is scored using the compression scoring function,18 defined as M–B–L, where M is the number of malignant findings for which the rule is true, B is the number of benign findings for which the rule is true, and L is the number of descriptors used in the rule. The best rule for each abnormality is the one that optimizes this scoring function. We limited the longest rule to a maximum of ten descriptors (L = 10).
ILP Algorithm
Similarly to the technique reported previously,1 we used Srinivasan's Aleph18 ILP system to learn rules from the set of examples. In this modified ILP analysis, we set Aleph to consider rules for each malignant finding in turn in the following manner: The search through the space of rules starts with the most general rule, based on information from the background knowledge, which is true for every mammographic finding whether benign or malignant. For each round, the best rule seen so far that meets the recall and precision settings above is selected and extended. Each descriptor from the selected finding is considered as an extension by adding the descriptor to the rule and calculating its score using the compression scoring function. This process of selecting the best rule seen so far and considering all possible extensions repeats until 10,000 rules have been considered for each malignant finding. The single best rule for each malignant finding is then reported. The algorithm was run on a single desktop machine with an Intel® Xeon® 2.33-GHz processor and 4 GB of RAM. The total time to run the algorithm on our dataset was approximately 8 h.
A total of 80 unique rules were learned that met the above criteria. Of all the rules, each rule was true, on average, for 40 malignant and 43 benign findings (precision of 48%) and contained five descriptors. Combining all the rules together, the entire set of 80 rules had a recall of 67% and a precision of 23% in the dataset of 62,219 abnormalities. A radiologist reviewed all 80 rules and identified potentially interesting rules based on known significant predictors of malignancy, such as spiculated margins and older age, a few of which appear in Table 2. Many of the descriptors used in the rules are already known to be predictive of malignancy. Similar to previous results, we found that high breast mass density frequently appeared in the set of rules. Of the 80 unique rules learned, 19 (24%) contained the high mass density descriptor.
Evaluation
After discovering that high mass density frequently appeared in rules learned by ILP, we evaluated the predictive ability of each descriptor alone and in comparison with other descriptors. We calculated the conditional probability of malignancy given each descriptor individually, which is defined as the fraction of findings that are both malignant and have the descriptor divided by the total number of findings that have the descriptor. Probabilities were smoothed using Laplacian smoothing to help mitigate the problem of rare descriptors. We then ordered the descriptors from the most predictive to the least predictive.
The top ten descriptors with their associated conditional probabilities are listed in Table 3. The majority of the descriptors contained within rules discovered by our ILP algorithm are already well-established predictors of malignancy; however, high mass density appears as the fifth most predictive indicator.
Knowledge Discovery Conclusions
The ILP method generated several rules that contained the high mass density descriptor, and shows that high mass density is a useful predictor of malignancy when used in conjunction with additional descriptors. Calculated conditional probabilities further confirmed this conclusion, demonstrating that high mass density is among the features most predictive of malignancy.
Validation of Knowledge Discovery
Dataset
In order to validate the conclusions generated by the data mining technique described above in an independent dataset, we collected data from 706 consecutive breast biopsies of masses performed between October 2005 and December 2007 at the University of Wisconsin Hospitals and Clinics Breast Center (a different medical center from the dataset of abnormalities described above). All female subjects who underwent image-guided core biopsy (ultrasound, stereotactic, or MRI-guided) for a solid, non-calcified mass seen on a diagnostic mammogram in which the mass density was described prospectively were eligible for this study. There were 180 masses that met these criteria. Each biopsy targeted a single mass.
Imaging and Evaluation
All mammographic studies were performed with dedicated mammography equipment, on either analog or digital machines. Of the 175 diagnostic mammograms performed, there were 69 (39.4%) analog mammographic examinations and 106 (60.6%) digital mammographic examinations. The type of equipment was chosen by the technologist based on availability. Eight interpreting radiologists participated in this study, all of which practice in the same academic group. Three of the radiologists are fellowship-trained in breast imaging.
Each radiologist described the mass prospectively as part of routine clinical practice at the time of imaging using the terminology of the fourth edition of the BI-RADS lexicon.7 Mass descriptors were used at the discretion of the interpreting radiologist and were entered directly into structured reporting software (PenRad Technologies®). Each mass was given a final BI-RADS assessment category by the interpreting radiologist.
Study End Points
After the mass was biopsied, a board-certified or board-eligible pathologist evaluated each sample to determine a final pathologic diagnosis. All pathology results were correlated with mammographic findings at a weekly radiologic pathology conference. Pathology results were grouped into benign and malignant categories based on the specific pathologic diagnosis. High-risk lesions, such as atypia or radial scar, were considered benign. For masses determined to be benign, we matched each subject to a hospital-based cancer registry and followed them for at least 12 months (mean 25.5, range 12–38).
Statistical Analysis
We collected the data by querying the structured reporting system for subject age, overall breast composition, all mass descriptors (density, shape, and margin), size of the mass, and image acquisition technique (digital or analog). We entered these data into a Microsoft Access Database and performed all statistical analyses using R, version 2.8.1 (R Foundation for Statistical Computing, Vienna, Austria). We grouped low and iso-dense masses together since a small proportion of the masses were considered low-density (16 of 180 masses, 8.9%). The log of the mass size was used in all analyses to account for outliers. We collapsed multi-level categorical variables (overall breast composition, mass shape, and mass margin) into dichotomous variables. For breast composition, the lower two categories (“almost entirely fat” and “scattered fibroglandular densities”) were compared to the higher two categories (“heterogeneously dense” and “extremely dense”). “Irregular” shape was compared to all other shape descriptors, and “spiculated” margin was compared to all other margin descriptors. Univariate analysis was performed to test the association between breast mass density, overall breast composition, image acquisition technique, age, and size of the mass and the probability of benign or malignant outcome. To account for multiple masses in the same subject, we used linear mixed-effect models to estimate p values, and bootstrapping to calculate the odds ratio. A p value of <0.05 was considered statistically significant for all analyses.
Logistic Regression
In order to evaluate the relative contribution of breast mass density to predicting malignancy while controlling for potentially confounding known predictors of malignancy, we created a logistic linear mixed effects model with benign or malignant outcome as the dependent variable. We used a backward stepwise variable selection method and determined the best fitting model based on the Akaike Information Criterion (AIC) and deviance differences. This type of model was used to address clinically important predictors and remove predictors from the model that did not improve prediction accuracy. Independent variables entered into the model included all collected variables listed above in addition to the random effect variable “interpreting radiologist.”
Results
The study population included 180 masses that were visualized on 175 diagnostic mammograms, among 173 subjects. Of the 180 masses, there were 109 benign masses and 71 malignant masses. Among the benign masses, 30 (27.5%) were high-density, while among the malignant masses, 51 (71.8%) were high-density. The results of the univariate analyses are presented in Table 4. Image acquisition technique was the only variable among those tested that was not found to be statistically significant.
In our logistic regression model, the mass descriptors high density (OR 6.6, CI 2.5–17.6), irregular shape (OR 10.0, CI 3.4–29.5), and spiculated margin (OR 20.4, CI 1.9–222.8) significantly predicted malignancy (Table 5). Although subject age (β = 0.09, p < 0.0001) also significantly predicted malignancy, the other independent variables tested did not improve prediction accuracy and were removed from the model.
Discussion
In this study, we confirm the conclusions of previous research by using ILP and calculated conditional probabilities on a dataset of structured mammographic findings. Both the ILP method and conditional probabilities showed that high mass density is a potentially important predictor of breast malignancy. Most importantly, we validated these conclusions in an independent dataset of mammographic findings collected prospectively during routine clinical practice.
One of the final steps in the knowledge discovery process is the evaluation of hypotheses generated using data mining techniques, which serves several important roles, including determining if the hypothesis is novel, medically important, and generalizable outside of the mined dataset. Although ILP has been used in the past1 to show that high mass density may be an important predictor of malignancy, that research only raised the possibility of the association but did not quantify results or test the conclusion in an independent dataset as we have done here. This step is critical for both verifying that the association is non-random or dataset-specific and proving its generalizability. In order to use the results from data mining techniques, validation analysis must be conducted to provide convincing evidence of the utility of the discovered knowledge.
The association between high mass density and malignancy is interesting because, although experts have asserted the association in the past,19 it has not been proven in the literature. The only study to evaluate this association concluded that the contribution of mass density to predicting malignancy was less than previously thought. Our study, which is larger and performed in a sample of consecutive biopsies collected prospectively, showed that mass density is an important adjunct descriptor that can be used to help stratify the risk of malignancy.
The conclusions of this study are clinically important for several reasons. First, in the setting of mammography, identifying novel predictors of breast cancer is important as the positive predictive value (PPV) of biopsy remains quite low (10–35%). By identifying additional features that aid in the diagnosis of cancer, we may be able to improve the PPV of biopsy. Second, our research shows the importance of clinical structured reporting and the collection of structured data. These data can be mined for the purpose of knowledge discovery and used for validation of discovered hypotheses. Finally, our study shows that we can take advantage of the computer's ability to survey large amounts of data to generate hypotheses, as well as the human's expertise in evaluating those hypotheses.
There were limitations to our study. First, although the datasets used in our study were from different institutions, they were collected in the same geographic region, thus making these datasets possibly more homogeneous than datasets from distinct geographic regions. Second, our method relies on the input of a radiologist to determine the importance of generated rules. In the future, we may be able to further automate the rule selection process using metrics, which would promote efficient rule selection. In this way, techniques such as ILP may help identify future research areas. For example, mammography data that include biopsy results could help determine the pathophysiologic factors that link high mass density with malignancy, which may lead to improvements in diagnosis and treatment. Still, we believe the results of this study are valid, and important conclusions can be drawn from this line of research.
Conclusion
The results of our study show that ILP with validation can be reliably used to help identify novel hypotheses in the large amounts of biomedical data available within the imaging enterprise. This kind of research is important to the future of imaging research, particularly in breast imaging, where the amount of available data becomes increasingly vast with the accelerating use of both ultrasound and magnetic resonance imaging. By using data mining techniques combined with targeted studies to validate hypotheses, we may improve the efficiency of discovering valuable medical knowledge.
References
Burnside ES, Davis J, Costa VS, et al: Knowledge discovery from structured mammography reports using inductive logic programming. AMIA Annu Symp Proc 96–100, 2005
Dang PA, Kalra MK, Blake MA, et al: Natural language processing using online analytic processing for assessing recommendations in radiology reports. J Am Coll Radiol 5(3):197–204, 2008
Lang M, Kirpekar N, Burkle T, Laumann S, Prokosch HU: Results from data mining in a radiology department: the relevance of data quality. Stud Health Technol Inform 129(Pt 1):576–580, 2007
Cios KJ, Teresinska A, Konieczna S, Potocka J, Sharma S: A knowledge discovery approach to diagnosing myocardial perfusion. IEEE Eng Med Biol Mag 19(4):17–25, 2000
Dzeroski S, Lavrac N: Introduction to inductive logic programming. In: Dzeroski S, Lavrac N Eds. Relational Data Mining. Berlin: Springer, 2001, pp. 48–71
Grinstead CM, Snell JL: Introduction to Probability, 2nd edition. Providence: American Mathematical Society, 1997
American College of Radiology: Breast Imaging Reporting and Data System (BI-RADS), 4th edition. Reston, VA, 2003
Baker JA, Kornguth PJ, Lo JY, Williford ME, Floyd Jr, CE: Breast cancer: prediction with artificial neural network based on BI-RADS® standardized lexicon. Radiology 196(3):817–822, 1995
Ciatto S, Cataliotti L, Distante V: Nonpalpable lesions detected with mammography: review of 512 consecutive cases. Radiology 165(1):99–102, 1987
Hall FM, Storella JM, Silverstone DZ, Wyshak G: Nonpalpable breast lesions: recommendations for biopsy based on suspicion of carcinoma at mammography. Radiology 167(2):353–358, 1988
Helvie MA, Pennes DR, Rebner M, Adler DD: Mammographic follow-up of low-suspicion lesions: compliance rate and diagnostic yield. Radiology 178(1):155–158, 1991
Lehman CD, Rutter CM, Eby PR, White E, Buist DS, Taplin SH: Lesion and patient characteristics associated with malignancy after a probably benign finding on community practice mammography. AJR Am J Roentgenol 190(2):511–515, 2008
Liberman L, Abramson AF, Squires FB, Glassman JR, Morris EA, Dershaw DD: The breast imaging reporting and data system: positive predictive value of mammographic features and final assessment categories. AJR Am J Roentgenol 171(1):35–40, 1998
Sickles EA: Nonpalpable, circumscribed, noncalcified solid breast masses: likelihood of malignancy based on lesion size and age of patient. Radiology 192(2):439–442, 1994
Varas X, Leborgne F, Leborgne JH: Nonpalpable, probably benign lesions: role of follow-up mammography. Radiology 184(2):409–414, 1992
Vizcaino I, Gadea L, Andreo L, et al: Short-term follow-up results in 795 nonpalpable probably benign lesions detected at screening mammography. Radiology 219(2):475–483, 2001
Jackson VP, Dines KA, Bassett LW, Gold RH, Reynolds HE: Diagnostic importance of the radiographic density of noncalcified breast masses: analysis of 91 lesions. AJR Am J Roentgenol 157(1):25–28, 1991
Srinivasan A: The Aleph Manual, 4th edition:http://www.comlab.ox.ac.uk/activities/machinelearning/Aleph/aleph_toc.html. Accessed May 24th, 2009.
Egan RL: Breast Imaging : Diagnosis and Morphology of Breast Diseases, Philadelphia: Saunders, 1988
Acknowledgements
This work was supported by the National Institutes of Health (grants K07-CA114181 and R01-CA127379).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Woods, R.W., Oliphant, L., Shinki, K. et al. Validation of Results from Knowledge Discovery: Mass Density as a Predictor of Breast Cancer. J Digit Imaging 23, 554–561 (2010). https://doi.org/10.1007/s10278-009-9235-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-009-9235-3