Abstract
A method that is easy to use, rapid, with a low cost of detecting viral nucleic acid in a biological sample represents the essential tool in targeted therapy. In this study, we report the use of paramagnetic microparticles covered by streptavidin and modified by an oligonucleotide probe with a specific viral sequence labeled by biotin to detect human immunodeficiency virus (HIV) and influenza virus subtype H5N1. The viral nucleic acids were primarily detected by adsorptive transfer stripping technique coupled with square wave voltammetry using carbon paste, hanging mercury drop or carbon nanotubes-based screen-printed working electrodes. Detection limits were estimated for both sequences down to picograms per 3 μl. To isolate the viral sequences, paramagnetic microparticles covered with biotin-labeled oligonucleotides were used. We calculated the yield of isolation for H5N1 and/or HIV sequences, which was defined as “isolated concentration of viral nucleic acid sequence”/“given viral nucleic acid sequence” × 100. We estimated the yield for both sequences as 59%. Moreover, we studied the influence of human serum, dsDNA and non-complementary sequence of nucleic acids on isolation of viral nucleic acids. We also used carbon nanotubes-based screen-printed electrodes coupled with micro-flow instrument to detect viral nucleic acids. We were able to isolate and detect nanogram amounts of nucleic acids.
Similar content being viewed by others
1 Introduction
Viral infections pose a threat to mankind (Gessain and Calattini 2008; McFee 2007). Certain viruses such as human immunodeficiency virus (HIV) or influenza viruses (mainly influenza A virus subtype H5N1) can evoke pandemics with high mortality. The H5N1 can cause illness in humans and many other animal species. A bird-adapted strain of H5N1 is the causative agent of H5N1 flu, commonly known as “avian influenza” or “bird flu”. The last great pandemics of influenza viruses were recorded in 1918, 1957 and 1968. Every time these pandemics were initiated by transformation of one of many influenza strains that freely circulated between wild and domestic birds to animal species, thereby a new, highly infectious (virulent) strain probably could be formed (Moscona 2008).
Human immunodeficiency virus is a retrovirus that can lead to acquired immunodeficiency syndrome (AIDS), a condition in humans in which the immune system begins to fail. HIV infection in humans is now pandemic. As of January 2006, the Joint United Nations Program on HIV/AIDS and the World Health Organization (WHO) estimate that AIDS has killed more than 25 million people since it was first recognized on 1 December 1981, making it one of the most destructive pandemics in recorded history. It is estimated that about 0.6% of the world’s population is infected with HIV. On the other hand, according to current estimates, HIV is set to infect 90 million people in Africa, which means that more than 15% of Africa’s population is infected (McGovern 2007).
Diagnostic possibilities are relatively limited. Nowadays they are focused on the direct cultural proof of identity or they result from the detection of viral antigen by the use of antibodies (Vernet 2007). The procedures utilizing molecular–biological methods, especially polymerase chain reaction, are also implemented (Neuzil et al. 2006a, b). To detect nucleic acids, electrochemical methods can be employed too. In the last decade, Professor Emil Palecek et al., published several reviews summarizing electrochemical research of nucleic acids (Fojta et al. 2008; Palecek 2002, 2008; Palecek and Jelen 2002). Professor Palecek is a pioneer in this field of research and his contribution to the understanding of electrochemical behavior of nucleic acids is unquestionable. Several major research centers targeting on electrochemical detection of nucleic acids and thus determining the direction of the research in this field have been established. Interest in electrochemical detection of nucleic acids clearly illustrates the increase in the number of papers according to Web of Science (Fig. 1). Due to using of electrochemically active labels the electrochemical detection can be considered as one of the most sensitive tools for detection of nucleic acids (Drummond et al. 2003; Gorodetsky et al. 2008; Kizek et al. 2002; Palecek 2004; Palecek and Jelen 2002; Palecek et al. 2002c; Teles and Fonseca 2008). However, the labeling of specific nucleic acids sequences is hardly to be done on site. Therefore, many scientists focus on direct nucleic acid detection (Jelen et al. 2004; Kourilova et al. 2005; Palecek 2002; Palecek et al. 2002a). Techniques based on simple and easy-to-use biological sample processing such as paramagnetic particles play an important role in this area (Palecek and Fojta 2007; Palecek et al. 2002b) as well as attempts to miniaturize a whole detection system e.g., screen-printed electrodes, which can be modified with various types of nano-materials (Huska et al. 2008b; Kumar and Chen 2008; Wang and Musameh 2004).
In this study, we report on the suggestion of a new method in the detection of viral nucleic acid based on paramagnetic microparticles (MPs). Particularly, we utilize MPs modified by streptavidin with high affinity to biotin for detection of influenza H5N1 and HIV.
2 Materials and methods
2.1 Chemicals and materials
All chemicals of ACS purity used and parafilm were purchased from Sigma-Aldrich Chemical Corp (USA) unless noted otherwise. Lyophilized highly polymerized DNA (Reanal, Hungary) was isolated from chicken erythrocytes (MW = 400,000 g/mol). Synthetic oligonucleotides, which were purified using high-performance liquid chromatography, were obtained from Generi Biotech (Czech Republic) with the following sequences: HIV: biotin-5′ CAA GCA GCC ATG CAA ATG TTA 3′ and complementary sequence: 3′ GTT CGT CGG TAC GTT TAC AAT 5′ (MW: 6900); H5N1: biotin-5′ CAA ACA GAT TAG TCC TTG CGA CTG 3′ and complementary sequence: 3′ GTT TGT CTA ATC AGG AAC GCT GAC 5′ (MW: 7363). Non-complementary sequence: 3′ GTA GGA CGA CGA TAC GGA GTA GA 5′. Stock standard solutions of the oligonucleotides (100 μg/ml) were prepared with water of ACS purity (Sigma-Aldrich) and stored in dark at −20°C. The concentration of oligonucleotides was determined spectrophotometrically at 260 nm using spectrometer Specord 210 (Analytic Jena, Germany). Deionized water underwent demineralization by reverse osmosis using the instrument Aqua Osmotic 02 (Aqua Osmotic, Czech Republic) and then it was subsequently purified using Millipore RG (Millipore Corp., USA, 18 MΏ)—MiliQ water. The pH value was measured using inoLab controlled by the personal computer program (MultiLab Pilot; Wissenschaftlich-Technische Werkstätten-WTW, Germany).
2.2 Instrumentation for viral oligonucleotide analysis
Isolation of viral oligonucleotides was carried out using paramagnetic microparticles covered by streptavidin and magnetic stand Dynal Magnetic Particle Concentrator-S both supplied by Dynal A.S (Norway). The diameter of the paramagnetic microparticles was 2.8 μm with binding capacity within the interval from 650 to 900 pmoles/mg beads. All experiments with paramagnetic microparticles were performed in RNA/DNA UV cleaner box Biosan (Latvia). For centrifuging and vortexing of a sample multispin MSC-3000 centrifuge (Biosan) placed in UV cleaner box was used. Denaturation was carried out at 85°C using the Thermomixer 5355 Comfort/Compact (Eppendorf, Germany). The buffers used in our experiments were as follows—(a) phosphate buffer I: 0.1 M NaCl + 0.05 M Na2HPO4 + 0.05 M NaH2PO4; (b) phosphate buffer II: 0.2 M NaCl + 0.1 M Na2HPO4 + 0.1 M NaH2PO4; (c) acetate buffer: 0.2 M CH3COOH + 0.2 M CH3COONa. Other details see in paper (Huska et al. 2009).
2.2.1 Hybridization procedure
The nucleic acid hybridization process was optimized previously (Huska et al. 2009). Briefly, the optimal composition of hybridization solution was as follows: 100 mM Na2HPO4 + 100 mM NaH2PO4, 0.5 M NaCl, 0.6 M Guanidium thiocyanate, 0.15 M Trizma base adjusted by HCl on pH of 7.5. The time of hybridization was 40 min and shaking 1,000 rpm (multispin MSC-3000 centrifuge (Biosan, Latvia).
2.3 Square wave voltammetric measurements
Square wave voltammetric measurements were performed with AUTOLAB PGS30 Analyzer (EcoChemie, Netherlands) connected to VA-Stand 663 (Metrohm, Switzerland), using a standard cell with three electrodes. A hanging mercury drop electrode (HMDE) with a drop area of 0.4 mm2, carbon paste electrode or carbon nanotubes-based screen-printed electrodes was employed as the working electrode. An Ag/AgCl/3 M KCl electrode served as the reference electrode. Glassy carbon electrode was used as the auxiliary electrode. For smoothing and baseline correction the software GPES 4.9 supplied by EcoChemie was employed. Square wave voltammetric (SWV) measurements were carried out in the presence of acetate buffer pH 4.6. SWV parameters: potential step 5 mV, frequency 280 Hz (Huska et al. 2009). The analyzed samples were deoxygenated prior to measurements by purging with argon (99.999%), saturated with water for 120 s.
2.3.1 Preparation of carbon paste electrode
The carbon paste (about 0.5 g, particles’ size 2–5 μm) was made of 70% graphite powder (Sigma-Aldrich) and 30% mineral oil (m/w) (Sigma-Aldrich; free of DNase, RNase and protease). This paste was housed in a Teflon body having a 2.5-mm-diameter disk surface. Prior to measurements, the electrode surface was renewed by polishing with a soft filter paper in preparation for measurement of a sample volume of 3 or 5 μl (Kizek et al. 2005; Masarik et al. 2003; Petrlova et al. 2007a, b).
2.3.2 Fabrication of carbon nanotubes-based screen-printed electrodes
Carbon nanotubes-based screen-printed electrodes. The sensor was fabricated by using a standard thick-film technology process with screen-printing semi-automat (UL 1505A, TESLA, CZ) and firing oven (TFF51, BTU International, USA) on an alumina substrate forming the three electrodes electrochemical sensor with dimensions of 25.4 × 5.0 × 0.33 (length × width × height) mm (Prasek et al. 2006). The ratio of the area of working electrode to the reference electrode 1/20 was used. The semidiameter of the working electrode was 0.4 mm and the internal semidiameter of the reference electrode was 0.6 mm and outer semidiameter 1.88 mm. Axial spacing of the contacts 1.2 mm, the width of the contact area of 0.5 mm and length of the contact area of 5 mm were used. The width of the inlet contacts for working and auxiliary electrodes was 0.5 mm and for reference electrode 0.3 mm. Power contacts have been isolated. Thick-film paste used for leads and contacts was AgPdPt based paste type ESL 9562-G (Electroscience, UK). The reference electrode (Ag/AgCl, ratio: 65:35) was printed from paste 5870 (DuPont, USA), the auxiliary electrode (Pt) from ESL 55445, dielectric layer from ESL 4913-G paste and working electrode (gold) form ESL 8881-B paste (all ESL ElectroScience, UK). All pastes were fired according to producer’s recommendations described in datasheets. The reference electrode was calibrated against saturated Ag/AgCl electrode (Prasek et al. 2006). The carbon multiwalled nanotubes (Sigma-Aldrich) were mixed with an organic binder, based on terpineol (Heraeus, Germany) and homogenized in mortar for 15 min. The printing was done using semi-automated screen-printer and dried at 150°C as described above.
2.4 Micro-flow system coupled with potentiostat
Micro-flow system consisted from peristaltic pump (Amersham Pharmacia Biotech, USA) connected with stock solution of mobile phase (acetate buffer, pH 4.6). The pump is connected to flow cell (BVT technologies, Czech Republic). Flow cell enables the use of AC1 and sensors in a flow through arrangement. The cell ensures the wall-jet flow around the working electrode and also contains the contact and output cable. The electrodes placed in the flow cell were connected to multi-mode potentiostat BioStat (ESA, Inc. USA). It is a four-channel system with three operating modes per channel (amps, volts, and temp). The system was connected through data bus USB to a personal computer. The homemade apparatus was connected to the first channel of the potentiostat. This apparatus consists of a basic plate on which the connector TX721 1115 with 2.54 pin spacing and the 0039532035 connector (Molex) with 1.25 mm pin spacing are placed. The connectors are designed for connection of two different screen-printed electrodes. For data processing, the software BioStat and MS Excel (Microsoft, USA) were used. The parameters of the analysis were as follows: channel number 1, sampling frequency 20 sps (samples/s), filter order 5. All electrochemical measurements were carried out in acetate buffer pH 4.6 at room temperature.
3 Results and discussion
Developing and designing on-chip paramagnetic-beads-based instruments for cell sorting, isolation of proteins, nucleic acids or other biologically active substances is a very intensively studied subject. Research is concentrated on fabrication of paramagnetic beads and also on design of the instrument itself (Estes et al. 2009). During the separation step it is necessary to ensure washing non-target molecules and the highest affinity to target molecules. Moreover, it is usually needed to avoid sedimentation of paramagnetic particles during the separation and washing cycles. The procedure that will ensure all these steps will make a significant contribution to improving the effectiveness of separation of molecules of interest (Kell et al. 2008; Le Drogoff et al. 2008; Lund-Olesen et al. 2006; Smistrup et al. 2006a, b, 2008; Xie et al. 2006).
3.1 Isolation and detection of viral nucleic acid
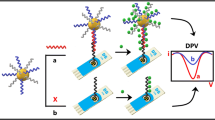
To detect viral nucleic acid probe coupling of paramagnetic microparticles (diameter 2.8 μm) and square wave voltammetry was employed. Oligonucleotide probe with specific viral-complementary sequence was labeled by biotin on 5′ tail. The vitamin (biotin) has a very high affinity to avidin/streptavidin (dissociation constant of 10−15 M), and this interaction has been utilized in many types of avidin-biotin technologies (Kizek et al. 2005; Kourilova et al. 2005; Masarik et al. 2003; Petrlova et al. 2007a, b). If paramagnetic microparticles are covered with streptavidin, they are able to bind oligonucleotide chains labeled with biotin. In our experiment magnetic microparticles modified with streptavidin were primarily covered with biotinylated oligonucleotides with complementary sequence to specific nucleic acids sequence of H5N1 or HIV (Fig. 1a). After rinsing steps (three times under shaking, Fig. 1b) microparticles covered by these probes interacted with certain viral nucleic acids sequence. Hybridization, rinsing and denaturation steps followed (Fig. 1c). The solution contained viral nucleic acids sequence only is electrochemically analyzed (Fig. 1d).
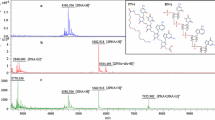
Electrochemical detection of viral nucleic acids (HIV and H5N1) was carried out at mercury (HMDE), carbon paste (CPE) and/or carbon nanotubes-based screen-printed (SPE) working electrodes. Due to small volumes of the obtained sample (10–30 μl) using adsorptive transfer stripping technique was needed, however, the sample volume can be lowered down to hundreds of nl (Adam et al. 2008). The principle of the procedure is based on the following steps: (a) renewing of a surface of a working electrode, (b) transferring of an electrode to a sample introduced on parafilm, (c) adsorbing of target molecule in a drop solution onto the surface at open circuit and/or superimposed potential, (d) rinsing the working electrode in a solution, and (e) transferring of the rinsed electrode to a supporting electrolyte and measuring (Fig. 2). Target nucleic acids’ sequences gave typical reduction signal of adenine and cytosine (peak CA) at −1.40 ± 0.02 V using HMDE. CA peaks were well developed and proportional to concentration of the sequence up to 1 μg/ml. The relative standard deviation of a measurement was 3%. The detection limits were estimated for both sequences down to 0.3 pg per 3 μl (Table 1). Using carbon electrodes (CPE and SPE) the sequences gave an oxidation signal of adenine at 1.30 ± 0.02 V for CPE and 1.15 ± 0.05 V for SPE. The relative standard deviation of a measurement was 6% for CPE and 11% for SPE. The enhancement of the deviations can be associated with difficult preparation of a similar surface of a working electrode (CPE) or with homemade fabrication of SPE. Moreover, the detection limits were 7 times higher (CPE) or even 15 times (SPE) as compared to HMDE. In spite of this fact, all the three tested working electrodes were still sensitive enough to detect viral nucleic acid in real samples (Huska et al. 2009).
a Onto paramagnetic microparticles with streptavidine modified surface biotinylated sequence of oligonucleotides complementary to specific viral sequence is bounded (a). To these microparticles viral nucleic acid is added (b). After hybridization microparticles with bounded viral nucleic acid are forced by magnet (c). Viral nucleic acid is released and electrochemically detected (d). b Principle of the adsorptive transfer stripping technique
3.2 Optimal hybridization parameters on magnetic microparticles surface
Previously we optimized the automatic procedure using paramagnetic microparticles to isolate adenine-containing oligonucleotides (Huska et al. 2008a, 2009). We followed the published procedure in our study. Briefly, the paramagnetic microparticles (10 μl) were pipetted with the solution in which they were kept in a test-tube. The test-tube was placed on a magnetic stand that separated the paramagnetic microparticles from the storage solution. The solution was then pipetted away in 20 μl of washing solution (phosphate buffer I). Further, the test-tube was moved from the magnetic stand and shaken in the hands so that the paramagnetic microparticles were equally dispersed in the washing solution. The washing and shaking processes were repeated six times. Following the rinsing steps, the paramagnetic microparticles were covered with biotin-labeled oligonucleotides. For this purpose, four concentrations of a probe (0, 5, 10 and 15 μg/ml) and two periods of modification (30 and 60 min) were tested. Following the modification, the particles covered with probes were washed prior to isolate viral nucleic acids (Fig. 1). The heights of signals of isolated viral nucleic acid are shown in Fig. 3. The signal obtained from analysis of viral nucleic acid isolated by paramagnetic microparticles covered by 5 μg/ml of the probe was higher when compared to the 15 μg/ml probes-modified microparticles. In conclusion, the higher concentration of the biotinylated probe probably resulted in the formation of overlapping layers on the microparticles’ surface, which negatively affected the hybridization process (Fig. 3). In addition, a 30 min long modification was enough for our purposes in spite of the fact that a 60 min long process resulted in an enhancement of the viral nucleic acids’ signal by more than 25% (not shown).
Isolation of viral nucleic acids oligonucleotides (10 μg/ml) by paramagnetic microparticles covered by biotin-labeled oligonucleotide (0, 5, 10 and 15 μg/ml). Detection was carried out at HMDE by using of adsorptive transfer stripping technique in connection with SWV. Volume of a sample 5 μl, time of accumulation 120 s
Further, the modified magnetic microparticles were prepared to test hybridization with the model viral nucleic acid sequence. The microparticles covered by biotin-labeled were kept in the presence of H5N1 and/or HIV sequences (total volume of the hybridization solution with the sample was 30 μl). Hybridization was proceeding on apparatus multispin MSC-3000 for the 40 min with shaking (1,400 rpm) at 25°C. Following hybridization, the paramagnetic microparticles were rinsed. The rinsed microparticles with the bound nucleic acid were placed into a thermomixer. The denaturation was carried out at 85°C for 5 min. Under heat treatment, viral nucleic acid was released from the microparticles. Following this step, the magnetic stand was used to force the paramagnetic microparticles, and the solution containing only the viral nucleic acid was transferred into a new test-tube. The solution was subsequently analyzed. Isolated molecules of nucleic acids provided at working electrodes used well-repeatable signals such as: (i) the characteristic redox signal of adenine and cytosine at HMDE and (ii) the oxidative signals of individual nuclei acid bases at CPE and/or SPE.
3.3 Detection of the presence of viral nucleic acid
In our following experiments we detected viral nucleic acids (given concentration of viral nucleic acid sequences 0.7(14), 1.3(26), 2.0(40), 2.7(54), 3.3(66) and 5.0(100) μg/ml (ng content of the sequence in 20 μl)) isolated by paramagnetic microparticles. The isolated nucleic acids gave well repeatable and similar signals at HMDE, CPE and SPE (Fig. 4). The possibilities of detecting nucleic acids at the carbon electrodes represent an important step in the construction of an easy to use on site electrochemical detector. The obtained signals were proportional to the given concentration of viral nucleic acids: HMDE HIV: y = 80.57x − 123.5, R 2 = 0.960; HMDE H5N1: y = 96.64x − 127.7, R 2 = 0.968; CPE HIV: y = 333.5x − 505, R 2 = 0.972; SPE HIV: y = 233.0x − 358.0, R 2 = 0.968; CPE H5N1: y = 333.3x − 475.8, R 2 = 0.982; SPE H5N1: y = 260.4x − 370.3, R 2 = 0.983. Moreover, we calculated the yield of the isolation for H5N1 and/or HIV sequences, which was defined as “isolated concentration of viral nucleic acid sequence”/”given viral nucleic acid sequence” × 100. We estimated the yield for both sequences as 59%. It is clear that less than half of given amount of nucleic acids is lost during isolation. On the other hand, the estimated yield is relative higher considering the type of sample, kind of molecule of interest and way of isolation (Palecek and Fojta 2007).
Detection of viral nucleic acids oligonucleotides by electrochemical sensors. Concentration of applied HIV and/or H5N1 ODN (0, 0.7, 1.3, 2.0, 2.7, 3.3 and 5.0 μg/ml) on paramagnetic microparticles covered by biotin-labeled probe oligonucleotide (5 μg/ml). Time of hybridization was 40 min, temperature 25°C, shaking and 1,400 rpm, total volume samples 20 μl. Nucleic acids detection at (a) HMDE and/or (b) CPE/SPE
3.4 Interference of biological sample
For rapid diagnostics, detection of viral nucleic acids directly in body fluids is needed. To test proposed methods we added viral nucleic acids to human serum sample obtained from healthy laboratory stuff and isolated it by paramagnetic microparticles covered by biotinylated oligonucleotides. It is shown in Fig. 5 that the presence of interfering substances in blood serum significantly influences electrochemical response. Using HMDE as working electrode the negative effect of interferences was more evident at both types of viral nucleic acids. The declining of the signal is sharp as it follows from slopes: HIV HMDE: y = −17.00x (R 2 = 0.97); H5N1 HMDE: y = −17.40x (R 2 = 0.87); HIV CPE: y = −10.57x (R 2 = 0.90); HIV SPE: y = −10.44x (R 2 = 0.98); H5N1 CPE: y = −15.52x (R 2 = 0.88) H5N1 SPE: y = −13.65x (R 2 = 0.94). This effect is caused by higher affinity of high-molecular compounds to electrode surface. On the other hand carbon electrode demonstrated minor changes in electrochemical signal of HIV with increasing concentration of blood serum. H5N1 virus isolated by using of paramagnetic microparticles and detected by AdTS SWV gave the similar response at the surface of CPE or SPE compared to HMDE. It can be concluded that nucleotide sequence plays very important role in detection, because it is probable that some of interfering compounds may bind specifically to this sequence. In the case that we kept the modified paramagnetic particles in the presence of a sample not-containing nucleic acid (negative control), we did not detect any signal corresponding to electrochemical signals of nucleic acids. Non-specific sequences of oligonucleotides (sequences non-complementary to sequence of nucleic acids anchored on the surface of modified particles) can be bound on the surface of our prepared particles under specific conditions. Therefore, we tested binding of one non-complementary sequence of oligonucleotides on the surface of the particles. Under the same given concentration of non-complementary sequence we determined signal with 10% height compared to complementary sequence. In the presence of genomic DNA we did not observe marked adverse effects on binding of viral sequence on the surface of particles. Moreover, we estimated the yield for both sequences as 45.2% (HIV) and/or 42.8% (H5N1).
Effect of the main interfering substances presented in blood serum on hybridization process of viral nucleic acid oligonucleotides (10 μg/ml) onto the surface of paramagnetic particles covered by biotin-labeled probe oligonucleotide. Blood serum additions were 2, 4, 6, 10, 20 and 40% into hybridization solution. Time of hybridization was 60 min, temperature 25°C, shaking 1,400 rpm, total volume samples 20 μl. Nucleic acids detection at (a) HMDE and/or (b) CPE/SPE
3.5 Detection of viral nucleic acids by using of micro-flow instrument coupled with miniaturized potentiostat and carbon nanotubes-based screen-printed electrodes
We also attempted to use carbon nanotubes-based screen-printed electrodes coupled with micro-flow instrument for detection of viral nucleic acids. The whole detection system consisted of peristaltic pump, potentiostat and screen-printed micro-electrodes (Fig. 6). Kjeang et al., also reported on new electrochemical velocimetry approach with direct electrical output (Kjeang et al. 2007). In this study, a sample was introduced onto working electrode where interaction with working electrode took place. The interaction was terminated by rinsing of the electrode by acetate buffer (pH 4.6) as mobile phase with flow rate 200 μl/min and volume of a cell was 20 μl. For baseline establishing 10 μl of the supporting electrolyte was introduced on the electrodes surface, then 2 μl of the sample was added and the changes of the current were determined. The electrode surface was mechanically polished with redistilled water and filter paper. Primarily we detected all the four nucleic acid bases (adenine, guanine, cytosine and thymine) in the presence of acetate buffer with pH 4.6 at potential from 0 to 1,500 mV per 50 mV steps. Typical hydrodynamic voltammograms are shown in Fig. 7. The observed changes relate to the oxidation of single bases. The peak of adenine enhanced sharply from 850 mV with a maximum about 1,000 mV. The guanine peak steadily increased from 450 to 650 mV. Recently, it was shown that cytosine and thymine were also electroactive at the carbon electrodes (Oliveira-Brett et al. 2004). The highest peaks for cytosine and thymine were detected at 1,300 and 1,100 mV, respectively. Moreover, the genomic DNA, both in the native and denatured states, was detected under the same conditions. In comparison to native DNA, the same concentration of the denatured state gave a higher signal of more than 50%. On the basis of the hydrodynamic voltammograms, we selected 1,000 mV for our following experiments, in which the change of the oxidation signal of the nucleic bases on their increasing concentration was determined. The calibration curves with correlation coefficient higher than 0.98 were obtained for the single bases. The quantification limits evaluated at the fabricated electrodes were 0.2 μM for cytosine, 1.5 μM for guanine, 0.8 μM for thymine and 1.5 μM for adenine. The quantification limit of native DNA was about 1.5 μg/ml and that of denatured DNA was 0.5 μg/ml.
Besides single bases and genomic DNA, we also detected viral nucleic acid sequences. The calibration curves are shown in Fig. 8a. The dependences were linear with the interval from 0.2 to 25 μg/ml (HIV: y = 44.59x + 10.75, R 2 = 0.994; H5N1: y = 36.06x − 5.651, R 2 = 0.996); the relative standard deviation of a measurement was 6%. Further, we isolated viral oligonucleotides by paramagnetic particles and then detected by above-described flow instrument. In simple test electrochemical responses of certain nucleic acids (0, 5, 10, 15 and 25 μg/ml) isolated by paramagnetic microparticles were determined (Fig. 8b). The height of peaks measured was proportional to given concentration as follows: HIV: y = 121.6x and H5N1: y = 95.0x.
Detection of viral nucleic acid by using of micro-flow instrument coupled with miniaturized potentiostat and screen-printed electrodes. In upper part of the figure scheme of miniaturized electrode is shown (R reference electrode, W working and A auxiliary electrode). (a) Dependence of HIV and H5N1 oligonucleotides’ peak height on concentration of viral nucleic acids (b) Change of peak height of viral nucleic acid (0, 5, 10, 15 and 25 μg/ml) measured at miniaturized electrode. Supporting electrolyte: 0.2 M acetate buffer, pH 4.6, flow rate: 200 μl/min, applied potential 1,000 mV
4 Conclusion
Human population is globally threatened by expansive pandemics caused by viruses such as human immunodeficiency virus and influenza viruses. Diagnostic possibilities are relatively limited. Here we propose methods based on streptavidin-modified paramagnetic microparticles to isolate viral nucleic acids. In conclusion, the particles coupled with electrochemical detection at the micro-electrodes can be considered to be one of the promising tools in the field of viral detection. In addition, it is shown that the proposed simple miniaturized detector coupled with micro-flow instruments might become the suitable basis for easy-to-use and rapid detection of viral nucleic acid.
References
Adam V, Baloun J, Fabrik I et al (2008) An electrochemical detection of metallothioneins at the zeptomole level in nanolitre volumes. Sensors 8:2293–2305
Drummond TG, Hill MG, Barton JK (2003) Electrochemical DNA sensors. Nat Biotechnol 21:1192–1199
Estes MD, Do J, Ahn CH (2009) On chip cell separator using magnetic bead-based enrichment and depletion of various surface markers. Biomed Microdevices 11:509–515
Fojta M, Jelen F, Havran L et al (2008) Electrochemical stripping techniques in analysis of nucleic acids and their constituents. Curr Anal Chem 4:250–262
Gessain A, Calattini S (2008) Emergence of simian foamy viruses in humans: facts and unanswered questions. Future Virol 3:71–81
Gorodetsky AA, Ebrahim A, Barton JK (2008) Electrical detection of TATA binding protein at DNA-modified microelectrodes. J Am Chem Soc 130:2924–2925
Huska D, Adam V, Trnkova L et al (2008a) Dependence of adenine isolation efficiency on the chain length evidenced using paramagnetic particles and voltammetry measurements. J Magn Magn Mater 321:1474–1477
Huska D, Hubalek J, Adam V et al (2008b) Miniaturized electrochemical detector as a tool for detection of DNA amplified by PCR. Electrophoresis 29:4964–4971
Huska D, Hubalek J, Adam V et al (2009) Automated nucleic acids isolation using paramagnetic microparticles coupled with electrochemical detection. Talanta (in press). doi:10.1016/j.talanta.2009.1004.1007
Jelen F, Kourilova A, Pecinka P et al (2004) Microanalysis of DNA by stripping transfer voltammetry. Bioelectrochemistry 63:249–252
Kell AJ, Stewart G, Ryan S et al (2008) Vancomycin-modified nanoparticles for efficient targeting and preconcentration of Gram-positive and Gram-negative bacteria. Acs Nano 2:1777–1788
Kizek R, Havran L, Fojta M et al (2002) Determination of nanogram quantities of osmium-labeled single stranded DNA by differential pulse stripping voltammetry. Bioelectrochemistry 55:119–121
Kizek R, Masarik M, Kramer KJ et al (2005) An analysis of avidin, biotin and their interaction at attomole levels by voltammetric and chromatographic techniques. Anal Bioanal Chem 381:1167–1178
Kjeang E, Roesch B, McKechnie J et al (2007) Integrated electrochemical velocimetry for microfluidic devices. Microfluid Nanofluid 3:403–416
Kourilova A, Babkina S, Cahova K et al (2005) DNA hybridization on membrane-modified carbon electrodes. Anal Lett 38:2493–2507
Kumar SA, Chen SM (2008) Electroanalysis of NADH using conducting and redox active polymer/carbon nanotubes modified electrodes—a review. Sensors 8:739–766
Le Drogoff B, Clime L, Veres T (2008) The influence of magnetic carrier size on the performance of microfluidic integrated micro-electromagnetic traps. Microfluid Nanofluid 5:373–381
Long GL, Winefordner JD (1983) Limit of detection. Anal Chem 55:A712–A724
Lund-Olesen T, Dufva M, Hansen MF (2006) Capture of DNA in microfluidic channel using magnetic beads: increasing capture efficiency with integrated microfluidic mixer. J Magn Magn Mater 311:396–400
Masarik M, Kizek R, Kramer KJ et al (2003) Application of avidin-biotin technology and adsorptive transfer stripping square-wave voltammetry for detection of DNA hybridization and avidin in transgenic avidin maize. Anal Chem 75:2663–2669
McFee RB (2007) Avian influenza: the next pandemic? DM-Dis-a-Mon 53:348–387
McGovern BH (2007) The epidemiology, natural history and prevention of hepatitis B: implications of HIV coinfection. Antivir Ther 12:H3–H13
Moscona A (2008) Medical management of influenza infection. Annu Rev Med 59:397–413
Neuzil P, Pipper J, Hsieh TM (2006a) Disposable real-time microPCR device: lab-on-a-chip at a low cost. Mol Biosyst 2:292–298
Neuzil P, Zhang CY, Pipper J et al (2006b) Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res 34:9
Oliveira-Brett AM, Piedade JAP, Silva LA et al (2004) Voltammetric determination of all DNA nucleotides. Anal Biochem 332:321–329
Palecek E (2002) Past, present and future of nucleic acids electrochemistry. Talanta 56:809–819
Palecek E (2004) Surface-attached molecular beacons light the way for DNA sequencing. Trends Biotechnol 22:55–58
Palecek E (2008) Fifty years of nucleic acid electrochemistry. Electroanalysis 21:239–251
Palecek E, Fojta M (2007) Magnetic beads as versatile tools for electrochemical DNA and protein biosensing. Talanta 74:276–290
Palecek E, Jelen F (2002) Electrochemistry of nucleic acids and development of DNA sensors. Crit Rev Anal Chem 32:261–270
Palecek E, Billova S, Havran L et al (2002a) DNA hybridization at microbeads with cathodic stripping voltammetric detection. Talanta 56:919–930
Palecek E, Fojta M, Jelen F (2002b) New approaches in the development of DNA sensors: hybridization and electrochemical detection of DNA and RNA at two different surfaces. Bioelectrochemistry 56:85–90
Palecek E, Kizek R, Havran L et al (2002c) Electrochemical enzyme-linked immunoassay in a DNA hybridization sensor. Anal Chim Acta 469:73–83
Petrlova J, Krizkova S, Supalkova V et al (2007a) The determination of avidin in genetically modified maize by voltammetric techniques. Plant Soil Environ 53:345–349
Petrlova J, Masarik M, Potesil D et al (2007b) Zeptomole detection of streptavidin using carbon paste electrode and square-wave voltammetry. Electroanalysis 19:1177–1182
Prasek J, Adamek M, Hubalek J et al (2006) New hydrodynamic electrochemical arrangement for cadmium ions detection using thick-film chemical sensor electrodes. Sensors 6:1498–1512
Smistrup K, Bruus H, Hansen MF (2006a) Towards a programmable magnetic bead microarray in a microfluidic channel. J Magn Magn Mater 311:409–415
Smistrup K, Tang PT, Hansen O et al (2006b) Micro electromagnet for magnetic manipulation in lab-on-a-chip systems. J Magn Magn Mater 300:418–426
Smistrup K, Bu MQ, Wolff A et al (2008) Theoretical analysis of a new, efficient microfluidic magnetic bead separator based on magnetic structures on multiple length scales. Microfluid Nanofluid 4:565–573
Teles FRR, Fonseca LR (2008) Trends in DNA biosensors. Talanta 77:606–623
Vernet G (2007) Use of molecular assays for the diagnosis of influenza. Expert Rev Anti-Infect Ther 5:89–104
Wang J, Musameh M (2004) Carbon nanotube screen-printed electrochemical sensors. Analyst 129:1–2
Xie X, Nie XR, Yu BB et al (2006) Rapid enrichment of leucocytes and genomic DNA from blood based on bifunctional core-shell magnetic nanoparticles. J Magn Magn Mater 311:416–420
Acknowledgment
The financial support from the grants KAN 208130801 and GACR 102/08/1546 is highly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adam, V., Huska, D., Hubalek, J. et al. Easy to use and rapid isolation and detection of a viral nucleic acid by using paramagnetic microparticles and carbon nanotubes-based screen-printed electrodes. Microfluid Nanofluid 8, 329–339 (2010). https://doi.org/10.1007/s10404-009-0464-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10404-009-0464-z