Abstract
The presentation of extracellular matrix (ECM) proteins provides an opportunity to instruct the phenotype and behavior of responsive cells. Decellularized cell-secreted matrix coatings (DM) represent a biomimetic culture surface that retains the complexity of the natural ECM. Microenvironmental culture conditions alter the composition of these matrices and ultimately the ability of DMs to direct cell fate. We employed a design of experiments (DOE) multivariable analysis approach to determine the effects and interactions of four variables (culture duration, cell seeding density, oxygen tension, and media supplementation) on the capacity of DMs to direct the osteogenic differentiation of human mesenchymal stem cells (hMSCs). DOE analysis revealed that matrices created with extended culture duration, ascorbate-2-phosphate supplementation, and in ambient oxygen tension exhibited significant correlations with enhanced hMSC differentiation. We validated the DOE model results using DMs predicted to have superior (DM1) or lesser (DM2) osteogenic potential for naïve hMSCs. Compared to cells on DM2, hMSCs cultured on DM1 expressed 2-fold higher osterix levels and deposited 3-fold more calcium over 3 weeks. Cells on DM1 coatings also exhibited greater proliferation and viability compared to DM2-coated substrates. This study demonstrates that DOE-based analysis is a powerful tool for optimizing engineered systems by identifying significant variables that have the greatest contribution to the target output.
Similar content being viewed by others
Introduction
The extracellular matrix (ECM) plays a pivotal role in regulating the maintenance and behavior of progenitor cells via physical interactions with cell surface proteins and the modulation of soluble growth factor concentrations within the cellular microenvironment.7,23 Thus, capitalizing on the innate ability of the ECM to modulate cell behavior has been of great interest in developing novel biomaterial surfaces that are better able to direct cell phenotype by recapitulating the in vivo cellular milieu.41,43 The design of cell culture interfaces that maintain cells in their native behavioral state or instruct their phenotypic transformation toward that of a desired tissue would be of great benefit in the advancement of cell-based therapies and tissue engineering. To date, this has been primarily pursued by the deposition of individual ECM proteins on a substrate surface.17,33,36 However, this approach fails to accurately mimic the complex protein composition within the endogenous ECM, an environment that can be efficiently generated by a number of cellular populations.
Mesenchymal stem cells (MSCs) represent a useful tool in the study of cell-secreted matrix coatings, as they produce generous amounts of ECM when cultured on tissue culture plastic (TCP) and other biomaterial surfaces. Upon decellularization of these cultures, the residual ECM coating retains the capacity to alter the phenotype of subsequently seeded cell populations.8,13,22 The composition of the in vivo ECM associated with each particular tissue has a profound effect on the phenotypic behavior of the engaged neighboring cells. Therefore, it is reasonable to expect that cell-secreted matrix coatings of differing composition will manipulate the properties of the adjacent cells during culture to varying degrees. The composition of ECMs deposited in vitro by progenitor cells is dependent upon the culture duration and microenvironmental culture conditions.1,4,20,30 Upon decellularization, ECM coatings deposited over different lengths of time and environmental conditions (e.g., oxygen tension, shear forces, soluble factors) will therefore acquire unique compositions that will determine their efficacy in modulating the phenotype of naïve stem cell populations.14,26 Hence, it is important to determine the ideal conditions under which to culture matrix-depositing MSCs prior to their decellularization for material-based therapeutic strategies.
The examination of interactions between large numbers of potentially contributing variables using so-called “One Factor At a Time” (OFAT) experimental approaches is ineffective due to the number of variable permutations and the likelihood of overlooking complex interactions of those variables. Alternatively, multivariable statistical analysis through a design of experiments (DOE) approach addresses these challenges by markedly reducing the number of variable combinations and repetitions to be examined, while simultaneously determining the significance of single input variables and combinations of such variables toward the ultimate response of the system. DOE represents a powerful tool that has previously been applied toward the optimization of developmental protocols from a broad array of scientific fields.9,28,40,42
Substrate-mediated cues directing human MSC (hMSC) fate toward the osteogenic lineage, such as those provided by the native ECM, offer a novel approach toward the development of biomaterial constructs designed to bridge and repair non-healing skeletal defects.5,11,29 We hypothesized that a DOE-based approach would provide an efficient means of engineering specific hMSC-secreted matrix coatings optimized to accelerate the osteogenic differentiation of naïve hMSCs. We first investigated this hypothesis by analyzing differences in the osteogenic differentiation and proliferation of naïve MSCs in the presence of MSC-secreted matrices engineered under distinct DOE-determined conditions. Upon characterizing cellular responses to these matrices, we validated DOE predictions of matrix efficacy at directing hMSC osteogenic fate with an in-depth analysis of the osteogenic capacity of two engineered hMSC-secreted matrix coatings.
Materials and Methods
Cell Culture
Human bone marrow-derived MSCs (hMSCs, Lonza, Walkersville, MD) were expanded without further characterization in alpha minimum essential medium (α-MEM, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (JR Scientific, Woodland, CA) and 1% penicillin and streptomycin (Mediatech, Manassas, VA). Cells were cultured under standard conditions and utilized at passages 3–5. Medium was further supplemented with 50 μg/mL ascorbate-2-phosphate (A2P) for one passage prior to experimental use to prime cells for enhanced matrix deposition.10 For studies examining the role of oxygen tension, the oxygen microenvironment was controlled as previously described using airtight chambers (Billups-Rothenberg, Del Mar, CA).16
DOE Model
A Box–Behnken experimental design was created with Design-Expert 8 software (Stat-Ease, Minneapolis, MN) to analyze the contribution of three continuous variables (seeding density, culture duration, and oxygen tension) and one discrete variable (media supplementation) toward the ability of hMSC-secreted matrices to direct hMSC osteogenic differentiation. Continuous variables were examined at low, medium, and high levels with a centrally repeated condition. Linear, quadratic, and two-factor interactions of each variable were assessed.
Preparation of Decellularized Matrices
hMSCs were cultured on 12-well plates under DOE-prescribed conditions with media changes performed every 3 days. Cells were seeded at densities from 2 × 104 to 8 × 104 cells/cm2 and allowed to attach overnight. Plates were then cultured for 3–15 days at oxygen tensions ranging from 5 to 21% in either supplemented media (SM: α-MEM + 50 μg/mL A2P) or osteogenic media (OM: α-MEM + 10 mM β-glycerophosphate, 50 μg/mL A2P, 10 nM dexamethasone). Wells were then decellularized in a manner similar to that previously described.8 Briefly, wells were rinsed with PBS and treated with 0.5% Triton X-100 (Sigma, St Louis, MO) in 20 mM NH4OH in PBS for 5 min at 37 °C. Wells were rinsed with PBS and treated with DNAse (Sigma, 200 units/mL PBS) for 1 h at 37 °C. Following additional PBS rinsing, plates were allowed to dry within a sterile biosafety cabinet for up to 12 h. Matrix-coated plates were stored at room temperature in the dark for up to 1 month prior to use. Plates coated with fibronectin (Sigma, 0.5 mL of 25 μg/mL solution for 2 h) and untreated TCP served as control substrates.
Characterization of Decellularized Matrices
Decellularization of 12-well plates was assessed via uptake of calcein AM (AnaSpec, Fremont, CA) and DNA quantification. Briefly, 1 mL of calcein solution (3 μg/mL in α-MEM) was added to cell layers both pre- and post-decellularization for 15 min at 37 °C. Calcein fluorescence was quantified using a microplate reader (BioTek, Winooski, VT) at 485/530 nm and imaged using a Nikon Eclipse TE2000-U fluorescent microscope. DNA content was quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA) and microplate reader as described.24
The morphology of hMSC-secreted ECMs was characterized by scanning electron microscopy after culture on Thermanox plastic coverslips (Nunc, Rochester, NY) over 2 weeks as described.39 Cytochemical staining of 12-well plates was performed by fixing cultures and decellularized matrices with 2% formaldehyde followed by rinsing in PBS. Plates were then stained for total protein with 0.1% coomassie brilliant blue in 80% H2O/20% methanol (MP, Solon, OH) for 15 min. Cell layers and decellularized matrices were also stained for glycosaminoglycan content. Wells were rinsed with 1% acetic acid followed by incubation for 15 min with 1% Alcian Blue 0.1 N HCl (Sigma).
Osteogenic Response of Naïve hMSCs
qPCR Analysis
hMSCs were seeded onto decellularized ECM-coated, fibronectin-coated, or uncoated TCP wells in SM at 7500 cells/cm2 and allowed to attach overnight. Culture medium was refreshed with OM the following day, and cells were cultured in standard conditions for 3–21 days. Total RNA was collected using the RNeasy Mini kit (Qiagen, Valencia, CA) and 250–500 ng of total RNA was then reverse-transcribed with the QuantiTect Reverse Transcription Kit (Qiagen). qPCR was performed using TaqMan1 Universal PCR Master Mix (Applied Biosystems, Foster City, CA) on a Mastercycler1 realplex2 (Eppendorf, Westbury, NY); primers and probes for BGLAP (Hs01587814_g1), COL1A1 (Hs01076777_m1), IBSP (Hs00173720_m1), MRPL13 (Hs00204173_m1), RUNX2 (Hs00231692_m1), and SP7 (Hs01866874_s1) were purchased from Applied Biosystems. Amplification conditions were 50 °C for 2 min, 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Quantitative PCR results were normalized to RPL13 transcript level to yield ∆Ct. Fold change in expression was then calculated using the formula 2∆Ct, and fold changes between experimental groups and control wells were calculated to yield ∆∆Ct.
Alkaline Phosphatase Activity and Calcium Deposition
hMSCs were seeded onto decellularized matrix-coated, fibronectin-coated, and uncoated TCP wells as described above. Intracellular alkaline phosphatase (ALP) activity was quantified and normalized to sample DNA content.15 Unseeded decellularized matrices were analyzed at each timepoint as a control to distinguish intracellular ALP from ALP deposited and retained within the matrix, and ALP results were normalized to these control values. The distribution of mineralized deposits on the ECMs was analyzed qualitatively by alizarin red staining, while calcium deposition at 0, 1, 3, and 5 weeks was analyzed quantitatively as previously described.15,24
Cell Proliferation, Viability, and Attachment
To assess the contribution of distinct ECMs toward cellular proliferation and viability, hMSCs were seeded onto decellularized matrix-coated, fibronectin-coated, and uncoated TCP wells in SM at 4000 cells/cm2, allowed to attach overnight, and cultured in OM as described above. Cell proliferation was measured by quantifying DNA concentration in each well at 1, 4, and 7 days. Cell viability was quantified by measuring metabolic activity with a 10% solution of alamarBlue (AbD Serotec, Raleigh, NC) for 3 h at 1, 4, and 7 days.24 In addition, cell viability was examined under stressful conditions. Briefly, hMSCs were seeded onto each experimental surface at 50,000 cells/cm2, allowed to attach overnight, and cultured in hypoxia chambers (1% O2) for 24 h with serum-free media. Cell viability was assayed via calcein uptake. To characterize the ability of cells to attach to each substrate, hMSCs were seeded at 50,000 cells/cm2 in SM and allowed to attach to culture surfaces for 1 or 4 h. Wells were then rinsed with PBS, and cell retention was quantified by calcein uptake.
Statistical Analysis
Data are presented as mean ± standard error unless otherwise stated. Statistical analysis was performed using paired Student’s t tests and one-way ANOVA where applicable. Statistical analysis of DOE experiments was performed by Design-Expert 8 software (Stat-Ease). p Values less than 0.05 were considered statistically significant.
Results
DOE Model
A DOE-based approach was utilized to examine the effect of culture conditions when producing hMSC-secreted decellularized matrices (DMs) to direct the osteogenic potential of naïve hMSCs. Four distinct culture variables (Table 1) were input into a Box–Behnken experimental design, resulting in 26 unique culture environments for creating distinct DMs. The osteogenic and proliferative responses of naïve hMSC to these DMs were determined by qPCR and DNA quantification and compared to that of hMSCs cultured on TCP alone following 7 days in OM. Each response output was found to be significantly influenced by one or more input variables (p < 0.05), and the linear, quadratic, and two-factor interactions of each variable were assessed (Table 2).
DOE predictions of the naïve hMSC response to DMs engineered using different culture conditions were generated based on experimental results (Fig. 1). mRNA expression of osterix (SP7), a zinc finger-containing transcription factor required for osteoblast differentiation and bone formation,35 increased 8- to 35-fold in naïve hMSCs cultured on DMs after 7 days compared with cells cultured on TCP alone (Figs. 1a and 1b). SP7 expression was significantly higher on DMs with extended culture durations and deposited in SM (Fig. 1a) rather than OM (Fig. 1b). mRNA expression of RUNX2, another key transcription factor associated with osteoblast differentiation,31 underwent a more modest 1- to 2-fold increase on DMs over TCP (Figs. 1c and 1d). RUNX2 expression was modulated in both a linear and quadratic fashion by the culture duration and oxygen tension under which DMs were deposited, with extended culture duration and higher oxygen tension each yielding increased RUNX2 expression by hMSCs. Expression of bone sialoprotein (IBSP) mRNA, generating a protein component of native bone ECM that enhances matrix mineralization,18 increased 2- to 10-fold in naïve hMSCs cultured on DMs compared with TCP, with increases in IBSP correlating with increased DM culture duration (Figs. 1e and 1f). Similar to SP7, IBSP was also present at significantly higher levels in naïve hMSCs cultured on DMs deposited in SM (Fig. 1e) compared with those deposited in OM (Fig. 1f). Finally, hMSC proliferation increased significantly when cultured on DMs deposited over extended time periods and in OM (data not shown). Culture conditions not specifically noted in Figs. 1a–h are constant at their median value.
DMs engineered using extended culture duration, higher seeding densities, ambient oxygen tension, and A2P supplemented media appeared most effective at driving naïve hMSC osteogenesis, as determined by 3D surface model graphs exhibiting the correlation between culture conditions during matrix deposition and the resulting response of naïve hMSCs. (a) SP7 (osterix) expression in SM (21% O2), (b) SP7 expression in OM (21% O2), (c) RUNX2 expression in SM, (d) RUNX2 expression in OM, (e) IBSP expression in SM, and (f) IBSP expression in OM at 7 days. Data are fold change over expression in hMSCs on TCP
Results from the DOE-based experiments indicated that hMSC-secreted matrices deposited over longer durations (15 days) with higher initial seeding densities (8 × 104 cells/cm2) under higher oxygen tensions (21% O2) and in SM produce the most effective osteogenic substrates after decellularization. As verification of each of these findings would prove cumbersome and to further explore the efficacy of this model, we performed in-depth analyses of two distinct DMs predicted to be most effective (DM1) or somewhat less effective (DM2) at instructing the osteogenic differentiation of naïve hMSCs. DM1 was deposited over 15 days in 21% O2, while DM2 was deposited over 6 days in 5% O2. Both DM1 and DM2 were produced using initial seeding densities of 8 × 104 cells/cm2 and cultured in SM.
Characterization of Decellularized Matrices (DMs)
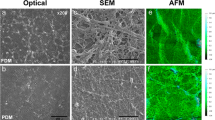
Decellularization of confluent hMSC layers (DM1) was confirmed by quantification of residual calcein uptake and DNA present within 12-well plates upon application of our decellularization protocol. Calcein uptake was virtually eliminated upon decellularization (Fig. 2a), with a 98.6% reduction in fluorescence observed in decellularized wells. Fluorescent microscopy images of calcein-treated wells before (Fig. 2c) and after decellularization (Fig. 2f) confirmed an apparent reduction in viable cells. Quantification of soluble DNA present within DM1-coated wells treated with cell lysis buffer also revealed a 99.9% reduction upon decellularization (Fig. 2b). Bright field and scanning electron microscopy were used to image DM1 hMSC cell layers before (Figs. 2d and 2e) and after decellularization (Figs. 2g and 2h), respectively. These techniques revealed a visible residual substrate composed in part of both proteins and glycosaminoglycans, as determined by staining with coomassie brilliant blue and Alcian blue (data not shown).
Quantitative analysis of the decellularization of DM1 utilizing (a) calcein uptake (n = 3) and (b) DNA quantification (n = 6). Fluorescent microscopy images of calcein uptake pre- (c) and post-decellularization (f). Bright field images (100× magnification) of cell layers pre- (d) and post-decellularization (g). SEM images of cell layers pre- (e) and post-decellularization (h) at 8172× magnification. Scale bars represent 250 μm (c, d, f, g) and 2 μm (e, h). *p < 0.0001 vs. pre-decell layers (a, b)
qPCR
We analyzed gene expression of five markers related to osteogenic differentiation (osterix, Runx2), as well as bone mineralization and ECM deposition (bone sialoprotein, osteocalcin, collagen 1a) over 21 days in hMSCs cultured on DM1, DM2, fibronectin, and TCP. Osterix expression from hMSCs cultured on DM1 was significantly higher than all other groups at 7 days, with a roughly 20-fold increase over hMSCs cultured on TCP, and a 2-fold increase over hMSCs cultured on DM2 (Fig. 3a). Both DM1 and DM2 were effective at stimulating osterix expression over the control surfaces at days 7, 11, and 15. Similar to prior DOE experimental results obtained during initial application, increases in RUNX2 expression resulting from hMSC culture on DM1 and DM2 were more muted. A roughly 2-fold increase was observed for cells on DM1 and DM2 over control groups at 7 days, with RUNX2 expression returning to those present in hMSCs cultured on TCP by 15 days (Fig. 3b).
DM1 enhanced expression of several ostegenic marker genes in hMSCs compared to DM2 or control substrates when probed by qPCR in hMSCs cultured on each substrate for 3–21 days: (a) SP7, (b) IBSP, (c) RUNX2, (d) BGLAP (osteocalcin), and (e) COL1A1. Data were normalized to MRPL13 transcript level and reported as fold change in mRNA expression vs. TCP controls. # p < 0.05 vs. all groups, † p < 0.05 vs. FN and TCP, *p < 0.05 vs. TCP only (n = 3)
Bone sialoprotein (IBSP) mRNA expression in hMSCs cultured on DM1 was significantly higher than all other groups from day 11 through 21, peaking with a roughly 75-fold increase over TCP controls at 11 days (Fig. 3c). hMSCs cultured on DM2 also showed a trend for increased IBSP expression over control groups from day 7 through 21. mRNA expression of osteocalcin (BGLAP), a common biochemical marker of bone formation,27 exhibited a significant increase in hMSCs cultured on DM1-coated wells over all other groups at days 3, 7, 15, and 21 (Fig. 3d). This expression peaked with a roughly 7-fold increase over TCP and 2-fold increase over DM2 at 7 days. Finally, collagen 1a expression, an essential building block of the major organic component of native bone ECM, was significantly increased in hMSCs cultured on DM1 compared with all other groups at 11 days (Fig. 3e).
Alkaline Phosphatase Activity and Calcium Deposition
Quantification of intracellular ALP activity in hMSCs cultured on DM1, DM2, fibronectin, and TCP in OM over 21 days revealed a significant increase within cells cultured on DM1 and DM2 at days 7 and 21 compared to controls (Fig. 4). No discernable differences in ALP levels were detectable between cells cultured on DM1 and DM2 coated plates with the exception of a reduction in ALP on DM1 coated plates on day 3.
Calcium deposition from hMSCs cultured on each substrate in OM was assayed both quantitatively and qualitatively over 5 weeks. Quantitative analysis revealed a significant increase in calcium deposition from hMSCs cultured on DM1 over all other groups at weeks 3 and 5, with a roughly 4-fold increase over control groups at week 3 (Fig. 5a). hMSCs cultured on DM2 also showed a significant increase in calcium deposition at week 5. No appreciable calcium deposition was noted after 1 week of hMSC culture, nor was any detectable calcium present in DM1 or DM2 prior to hMSC seeding. Qualitative analysis of wells via alizarin red staining also revealed a significant increase in calcium deposition by hMSCs cultured on DM1 (Fig. 5b) over all other groups (Figs. 5c–e) by 3 weeks.
hMSCs cultured on DM1 exhibited increased calcium deposition compared to cells on DM2 or control substrates. (a) Calcium deposited from hMSCs cultured on each substrate for 1, 3, and 5 weeks. Alizarin red staining of fixed hMSC layers cultured for 3 weeks on (b) DM1, (c) DM2, (d) FN, and (e) TCP. # p < 0.05 vs. all groups, † p < 0.05 vs. FN and TCP (n = 3–4)
Cell Proliferation, Viability, and Attachment
We detected a significant increase in the proliferative potential of hMSCs cultured on DM1 over all other groups at days 4 and 7 (Fig. 6a). hMSCs cultured on DM2-coated wells also proliferated faster than cells cultured on control surfaces. Furthermore, we observed differences in cell viability and metabolism of hMSCs cultured on the four surfaces. We detected a significant increase in alamarBlue reduction, an indicator of cell metabolism, from hMSCs cultured on DM1-coated wells over cells cultured on DM2-coated wells and TCP at all timepoints (Fig. 6b). hMSCs cultured on DM2-coated wells also showed a slight but significant increase in alamarBlue reduction at days 1 and 7 in comparison to hMSCs cultured on TCP alone. The ability of each of the four substrates to influence cell viability under harsh environmental conditions, such as those faced upon implantation in vivo, was also examined by total calcein uptake within wells following 24 h in a serum-free and hypoxic environment. We observed significantly greater calcein uptake by hMSCs cultured on DM1-coated wells compared to cells cultured in control TCP wells (Fig. 6c).
hMSC proliferation and viability are enhanced when cultured on DM1 compared to culture on DM2 or control substrates. (a) Total DNA on each substrate at 1, 4, and 7 days post-seeding. (b) AlamarBlue reduction by hMSCs cultured on each substrate at 1, 4, and 7 days. (c) Calcein uptake by hMSCs seeded on each substrate and under environmental stress for 24 h. # p < 0.05 vs. all groups, ‡ p < 0.05 vs. DM2 and TCP, † p < 0.05 vs. FN and TCP, *p < 0.05 vs. TCP only (n = 4–6)
The capacity of hMSCs to attach to DM1, DM2, fibronectin, and TCP substrates was assayed by calcein uptake within the wells at 1 and 4 h post-seeding. After 1 h, only fibronectin-coated wells exhibited a significant increase in calcein uptake. However, we detected significant increases in calcein uptake 4 h post-seeding for cells on both DM1- and DM2-coated wells compared with TCP wells, and calcein uptake in DM2-coated wells was increased over fibronectin-coated wells (Fig. 7).
Discussion
The successful design and implementation of constructs that mimic the properties of host tissue represents a fundamental goal of tissue engineering and regenerative medicine. Along with the types of cells present, the ECM is a defining component of each tissue in the body and therefore should be considered in the design of culture surfaces and tissue engineered constructs. Common approaches toward ECM incorporation in biomaterial design include (1) the coating of material surfaces with recombinant ECM proteins and peptides1,37 and (2) the use of decellularized allogenic or xenogenic tissue scaffolding to bridge the tissue defect and influence cellular behavior.2,19 However, the presentation of individual matrix proteins fails to capture the complex composition and architecture found within native ECM, and decellularized tissues suffer from immunological concerns while also lacking the reproducibility, availability, and tailorability available with synthetic materials.3,7,12 Cell-secreted matrix coatings, such as those studied herein, offer the complexity of a biologically relevant ECM, while enabling the potential synergism of an underlying, highly tunable synthetic backbone.
hMSCs represent a valuable tool for testing our hypothesis due to their multipotent nature and secretion of large quantities of matrix in culture, presumably depositing transitional matrices associated with the induced lineages along the way. As the complexity of environmental variables influencing the composition of the hMSC-secreted matrix is not fully understood, recent studies analyzing the ability of DMs to influence naïve MSC differentiation have focused on determining the relevance of various parameters including culture duration and the presence of soluble osteogenic cues.26,32 In order to minimize the number of experiments necessary to correlate such conditions with DM efficacy to modulate cell phenotype while maximizing experimental returns, we employed a DOE approach to analyze four conditions associated with hMSC-matrix deposition. This strategy is invaluable when a full knowledge of input variables and their effect on the system as a whole is poorly understood. A response surface method approach was employed to observe the curvature of our system response as a result of variable changes, and a Box–Behnken 3-level factorial design was chosen in lieu of a typical 5-level central composite design (CCD), as the more numerous and axial variable levels associated with the CCD are more difficult to carry out with biological systems. In this instance, a DOE-based approach allowed for more than a 2-fold reduction in the number of experimental groups and 5-fold reduction in the number of experimental samples necessary to perform a complementary, full-factorial designed experiment (n = 3). In the case of 5 or more variable systems, the reduction in the number of experimental groups becomes even more pronounced, allowing for faster optimization of engineered systems, and in turn, the isolation of those system variables which have the greatest impact and merit further study.
The DOE-based approach in our study revealed unanticipated correlations between conditions under which hMSC-deposited decellularized matrix coatings are created and their resulting capacity to direct the osteogenic differentiation of naïve hMSCs. Previous studies have examined links between the stage of hMSC differentiation prior to decellularization and the ability of the resulting DMs to drive osteogenesis. For example, Hoshiba et al. 26 demonstrated that an early stage transitional osteogenic matrix deposited by hMSCs in 2D was most efficient, while Liao et al. 32 observed that a more mature mineralized matrix coating was superior in a 3D murine cell model. Our data indicate that DMs deposited by hMSCs cultured in media supplemented with A2P only, and therefore in a comparably undifferentiated state, are more effective at driving hMSC osteogenesis than those DMs deposited by cells exposed to standard osteogenic media.
Mineralization of the DM also appeared unnecessary to drive naïve cell differentiation, as both DM1 and DM2 did not contain detectable levels of calcium prior to decellularization yet enhanced osteogenic differentiation compared to TCP. However, we did observe that culture duration of hMSCs for up to 15 days facilitated greater matrix deposition and more effective osteogenic DMs. Matrix deposition beyond 15 days generally resulted in curling at the edges of the cell layers, producing DMs that failed to completely adhere to the culture dish and did not fully cover the culture area. While no significant two-factor interactions were detected in our study, a trend was noted between culture duration and media type in driving osterix expression in naïve hMSCs (p = 0.089). The capacity of culture duration to drive osterix expression was more pronounced in supplemented media (Fig. 3a) than osteogenic media (Fig. 3b).
While the results of our study revealed several novel correlations between DM design and resultant hMSC phenotype, these findings should not be taken as rigid guidelines by which all osteogenic DMs should be engineered. Instead, these findings serve as a proof-of-principle that a DOE-based approach is a useful tool in optimizing unique biologically engineered systems. Differences in our results from previously published studies such as those pertaining to matrix mineralization may be due in part to disparities in the material surface, dimensionality, cell source, cell passage number, decellularization technique, and media type utilized in each study, all of which could affect DM composition and the outcome of a DOE-based optimization study.
To further probe the results from our DOE model, we characterized the osteogenic potential of the DOE-determined optimal matrix (DM1), as well as a second matrix (DM2) whose osteogenic potential was predicted to fall between that of DM1 and our control substrate TCP. Fibronectin-coated TCP also served as a single ECM protein-coated control, as fibronectin has been repeatedly described to influence adhesion, proliferation, and differentiation of numerous cultured cell populations.25 Although both DMs were more effective osteogenic substrates when compared to control surfaces, DM1 significantly outperformed DM2 as indicated by increases in the expression of multiple osteogenic genes, accelerated calcium deposition, and enhanced proliferative potential. These data show the power and accuracy of the DOE-based experimental approach to quickly isolate effective conditions that optimize multivariable input systems.
The assays and methods chosen in our study were selected in an attempt to effectively determine the impact of our substrates on naïve hMSC phenotype. To quantify osteogenic differentiation, we utilized qPCR in addition to ALP and calcium quantification. ALP and calcium are commonly sequestered within decellularized matrices, thereby making it difficult to discern whether measurements are indicative of the newly seeded cells alone or a combination of the cells and matrix. To address this challenge, we maintained unseeded control DMs at each time point and subtracted ALP quantities detected in those matrices from hMSC-seeded DMs. While our qPCR and calcium data confirmed the impressive osteogenic potential of our DMs, ALP activity was only slightly higher in hMSCs cultured on DM1 and DM2 compared to controls, possibly due to feedback inhibition from extracellular ALP present in the scaffolds. This problem was rendered mute in our calcium quantification assays, as the DMs studied contained no appreciable calcium prior to seeding with naïve hMSCs.
qPCR results from our study and previous work38 demonstrate that cell-secreted matrix coatings have the capacity to alter gene expression within progenitor cell populations. However, the method or methods by which this occurs are still unclear. Grunert et al. demonstrated that glycosaminoglycans present in DMs bind and modulate the efficacy of endogenous inductive factors such as bone morphogenetic protein-2 to direct hMSC osteogenic fate.22 Growth factors deposited by cells within the matrix prior to decellularization may therefore play a major role in determining DM efficacy, with naïve hMSCs likely secreting a unique array of trophic factors in comparison to cells found in a transitionally differentiated state.6,24 hMSC integrin binding ECM protein motifs may also contribute to the DM capacity to instruct cell phenotype. Previous reports confirm that the decellularization techniques utilized in our study retain key osteogenic ECM components upon cell removal,8 and MSC–ECM protein interactions have been significantly linked to both MSC potential for in vitro osteogenic differentiation34 and the pathway by which differentiation occurs.29 The capacity of osteogenic supplements to influence naïve hMSC differentiation may also be modulated by DM cell–substrate interactions, as dexamethasone may not be necessary for the osteogenic differentiation of MSCs cultured in mineralized matrix-coated constructs.32 To date, the mechanism of action for cell-secreted ECMs to direct cell fate, particularly with regard to ECM composition, is unclear. The presence of other components such as residual cell debris or intracellular proteins that may elicit an eventual immune response should be investigated more fully.21 While further characterization of osteogenic DMs and their interactions with progenitor cell populations is undoubtedly needed to fully unlock their mechanism of impact, our novel approach toward eliciting optimal cell-secreted DM design may help to better identify the specific DMs which merit such further study.
Conclusions
This study presents a simple but powerful DOE-based approach for engineering cell-secreted matrix coatings that can effectively drive the differentiation of progenitor cell populations toward a desired phenotype. This 2D proof-of-principle may serve as an effective tool toward the development of novel matrix-coated culture surfaces that “instruct” cell phenotype or for elucidating ECM-mediated pathways in stem cell differentiation. In addition, the translation of these techniques to a 3D model system could potentially allow for the design of synthetic/natural hybrid biomaterial constructs capable of modulating progenitor cell behavior following implantation within a tissue defect site.
References
Aulin, C., F. Foroughi, R. Brown, and J. Hilborn. Extracellular matrix-polymer hybrid materials produced in a pulsed-flow bioreactor system. J. Tissue Eng. Regen. Med. 3:188–195, 2009.
Badylak, S. F., D. O. Freytes, and T. W. Gilbert. Extracellular matrix as a biological scaffold material: structure and function. Acta Biomater. 5:1–13, 2009.
Badylak, S. F., and T. W. Gilbert. Immune response to biologic scaffold materials. Semin. Immunol. 20:109–116, 2008.
Bancroft, G. N., V. I. Sikavitsas, J. van den Dolder, T. L. Sheffield, C. G. Ambrose, J. A. Jansen, and A. G. Mikos. Fluid flow increases mineralized matrix deposition in 3D perfusion culture of marrow stromal osteoblasts in a dose-dependent manner. Proc. Natl. Acad. Sci. USA 99:12600–12605, 2002.
Bennett, K. P., C. Bergeron, E. Acar, R. F. Klees, S. L. Vandenberg, B. Yener, and G. E. Plopper. Proteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells. BMC Genomics 8:380, 2007.
Caplan, A. I., and J. E. Dennis. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 98:1076–1084, 2006.
Chen, X. D. Extracellular matrix provides an optimal niche for the maintenance and propagation of mesenchymal stem cells. Birth Defects Res. C 90:45–54, 2010.
Chen, X. D., V. Dusevich, J. Q. Feng, S. C. Manolagas, and R. L. Jilka. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J. Bone Miner. Res. 22:1943–1956, 2007.
Chen, W. L., M. Likhitpanichkul, A. Ho, and C. A. Simmons. Integration of statistical modeling and high-content microscopy to systematically investigate cell–substrate interactions. Biomaterials 31:2489–2497, 2010.
Choi, K. M., Y. K. Seo, H. H. Yoon, K. Y. Song, S. Y. Kwon, H. S. Lee, and J. K. Park. Effect of ascorbic acid on bone marrow-derived mesenchymal stem cell proliferation and differentiation. J. Biosci. Bioeng. 105:586–594, 2008.
Cool, S. M., and V. Nurcombe. Substrate induction of osteogenesis from marrow-derived mesenchymal precursors. Stem Cells Dev. 14:632–642, 2005.
Dahl, S. L., J. Koh, V. Prabhakar, and L. E. Niklason. Decellularized native and engineered arterial scaffolds for transplantation. Cell Transpl. 12:659–666, 2003.
Datta, N., H. L. Holtorf, V. I. Sikavitsas, J. A. Jansen, and A. G. Mikos. Effect of bone extracellular matrix synthesized in vitro on the osteoblastic differentiation of marrow stromal cells. Biomaterials 26:971–977, 2005.
Datta, N., Q. P. Pham, U. Sharma, V. I. Sikavitsas, J. A. Jansen, and A. G. Mikos. In vitro generated extracellular matrix and fluid shear stress synergistically enhance 3D osteoblastic differentiation. Proc. Natl. Acad. Sci. USA 103:2488–2493, 2006.
Davis, H. E., R. R. Rao, J. He, and J. K. Leach. Biomimetic scaffolds fabricated from apatite-coated polymer microspheres. J. Biomed. Mater. Res. 90:1021–1031, 2009.
Decaris, M. L., C. I. Lee, M. C. Yoder, A. F. Tarantal, and J. K. Leach. Influence of the oxygen microenvironment on the proangiogenic potential of human endothelial colony forming cells. Angiogenesis 12(4):303–311, 2009.
Dennis, J. E., S. E. Haynesworth, R. G. Young, and A. I. Caplan. Osteogenesis in marrow-derived mesenchymal cell porous ceramic composites transplanted subcutaneously: effect of fibronectin and laminin on cell retention and rate of osteogenic expression. Cell Transpl. 1:23–32, 1992.
Gordon, J. A., C. E. Tye, A. V. Sampaio, T. M. Underhill, G. K. Hunter, and H. A. Goldberg. Bone sialoprotein expression enhances osteoblast differentiation and matrix mineralization in vitro. Bone 41:462–473, 2007.
Grayson, W. L., S. Bhumiratana, C. Cannizzaro, P. H. Chao, D. P. Lennon, A. I. Caplan, and G. Vunjak-Novakovic. Effects of initial seeding density and fluid perfusion rate on formation of tissue-engineered bone. Tissue Eng. 14:1809–1820, 2008.
Grayson, W. L., F. Zhao, B. Bunnell, and T. Ma. Hypoxia enhances proliferation and tissue formation of human mesenchymal stem cells. Biochem. Biophys. Res. Commun. 358:948–953, 2007.
Griffiths, L. G., L. H. Choe, K. F. Reardon, S. W. Dow, and E. Christopher Orton. Immunoproteomic identification of bovine pericardium xenoantigens. Biomaterials 29:3514–3520, 2008.
Grunert, M., C. Dombrowski, M. Sadasivam, K. Manton, S. M. Cool, and V. Nurcombe. Isolation of a native osteoblast matrix with a specific affinity for BMP2. J. Mol. Histol. 38:393–404, 2007.
Guilak, F., D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke, and C. S. Chen. Control of stem cell fate by physical interactions with the extracellular matrix. Cell Stem Cell 5:17–26, 2009.
He, J., D. C. Genetos, C. E. Yellowley, and J. K. Leach. Oxygen tension differentially influences osteogenic differentiation of human adipose stem cells in 2D and 3D cultures. J. Cell. Biochem. 110:87–96, 2010.
Hidalgo-Bastida, L. A., and S. H. Cartmell. Mesenchymal stem cells, osteoblasts and extracellular matrix proteins: enhancing cell adhesion and differentiation for bone tissue engineering. Tissue Eng. B 16:405–412, 2010.
Hoshiba, T., N. Kawazoe, T. Tateishi, and G. Chen. Development of stepwise osteogenesis-mimicking matrices for the regulation of mesenchymal stem cell functions. J. Biol. Chem. 284:31164–31173, 2009.
Jaiswal, N., S. E. Haynesworth, A. I. Caplan, and S. P. Bruder. Osteogenic differentiation of purified, culture-expanded human mesenchymal stem cells in vitro. J. Cell. Biochem. 64:295–312, 1997.
Kim, S. H., and G. M. Lee. Development of serum-free medium supplemented with hydrolysates for the production of therapeutic antibodies in CHO cell cultures using design of experiments. Appl. Microbiol. Biotechnol. 83:639–648, 2009.
Kundu, A. K., and A. J. Putnam. Vitronectin and collagen I differentially regulate osteogenesis in mesenchymal stem cells. Biochem. Biophys. Res. Commun. 347:347–357, 2006.
Lecanda, F., L. V. Avioli, and S. L. Cheng. Regulation of bone matrix protein expression and induction of differentiation of human osteoblasts and human bone marrow stromal cells by bone morphogenetic protein-2. J. Cell. Biochem. 67:386–396, 1997.
Lian, J. B., A. Javed, S. K. Zaidi, C. Lengner, M. Montecino, A. J. van Wijnen, J. L. Stein, and G. S. Stein. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14:1–41, 2004.
Liao, J., X. Guo, D. Nelson, F. K. Kasper, and A. G. Mikos. Modulation of osteogenic properties of biodegradable polymer/extracellular matrix scaffolds generated with a flow perfusion bioreactor. Acta Biomater. 6:2386–2393, 2010.
Mauney, J. R., D. L. Kaplan, and V. Volloch. Matrix-mediated retention of osteogenic differentiation potential by human adult bone marrow stromal cells during ex vivo expansion. Biomaterials 25:3233–3243, 2004.
Mauney, J. R., C. Kirker-Head, L. Abrahamson, G. Gronowicz, V. Volloch, and D. L. Kaplan. Matrix-mediated retention of in vitro osteogenic differentiation potential and in vivo bone-forming capacity by human adult bone marrow-derived mesenchymal stem cells during ex vivo expansion. J. Biomed. Mater. Res. 79:464–475, 2006.
Nakashima, K., X. Zhou, G. Kunkel, Z. Zhang, J. M. Deng, R. R. Behringer, and B. de Crombrugghe. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17–29, 2002.
Ogura, N., M. Kawada, W. J. Chang, Q. Zhang, S. Y. Lee, T. Kondoh, and Y. Abiko. Differentiation of the human mesenchymal stem cells derived from bone marrow and enhancement of cell attachment by fibronectin. J. Oral Sci. 46:207–213, 2004.
Petrie, T. A., J. E. Raynor, D. W. Dumbauld, T. T. Lee, S. Jagtap, K. L. Templeman, D. M. Collard, and A. J. Garcia. Multivalent integrin-specific ligands enhance tissue healing and biomaterial integration. Sci. Transl. Med. 2:45–60, 2010.
Pham, Q. P., F. K. Kasper, L. Scott Baggett, R. M. Raphael, J. A. Jansen, and A. G. Mikos. The influence of an in vitro generated bone-like extracellular matrix on osteoblastic gene expression of marrow stromal cells. Biomaterials 29:2729–2739, 2008.
Rao, R. R., J. He, and J. K. Leach. Biomineralized composite substrates increase gene expression with nonviral delivery. J. Biomed. Mater. Res. 94:344–354, 2010.
Shah, M., and K. Pathak. Development and statistical optimization of solid lipid nanoparticles of simvastatin by using 2(3) full-factorial design. AAPS PharmSciTech. 11:489–496, 2010.
Singelyn, J. M., J. A. DeQuach, S. B. Seif-Naraghi, R. B. Littlefield, P. J. Schup-Magoffin, and K. L. Christman. Naturally derived myocardial matrix as an injectable scaffold for cardiac tissue engineering. Biomaterials 30:5409–5416, 2009.
Zahed, M. A., H. A. Aziz, M. H. Isa, L. Mohajeri, and S. Mohajeri. Optimal conditions for bioremediation of oily seawater. Bioresour. Technol. 101(24):9455–9460, 2010.
Zhang, Y., Y. He, S. Bharadwaj, N. Hammam, K. Carnagey, R. Myers, A. Atala, and M. Van Dyke. Tissue-specific extracellular matrix coatings for the promotion of cell proliferation and maintenance of cell phenotype. Biomaterials 30:4021–4028, 2009.
Acknowledgments
The authors are grateful for financial support from The Hartwell Foundation (to J.K.L.) and the California Institute for Regenerative Medicine UC Davis Stem Cell Training Program (CIRM T1-00006, CIRM TG2-01163).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Angelique Louie oversaw the review of this article.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Decaris, M.L., Leach, J.K. Design of Experiments Approach to Engineer Cell-Secreted Matrices for Directing Osteogenic Differentiation. Ann Biomed Eng 39, 1174–1185 (2011). https://doi.org/10.1007/s10439-010-0217-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-010-0217-x