Abstract
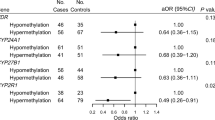
Dietary intake of one-carbon nutrients (methyl donors) and germline variants in the one-carbon metabolism genes may influence global DNA methylation level and methylation in promoter CpG islands. In this study, we evaluated the relationship between single nucleotide polymorphisms (SNPs) in the one-carbon metabolism pathway and DNA methylation status in colorectal cancer. Utilizing 182 colorectal cancers cases in two prospective cohort studies, we determined the CpG island methylator phenotype (CIMP) status on eight CIMP-specific promoters and measured LINE-1 methylation level that correlates well with genome-wide DNA methylation level. We genotyped 23 nonsynonymous SNPs in the one-carbon metabolism genes using buffy coat DNA. Most of the 23 SNPs in the one-carbon metabolism pathway were not significantly associated with CIMP-high status (≥6/8 methylated promoters). However, the MTHFR 429 Ala/Ala variant (rs1801131) and the TCN2 259 Arg/Arg variant (rs1801198) were associated with CIMP-high status (MTHFR 429 multivariate odds ratio (MV OR) = 7.56; 95% confidence interval (CI), 1.32–43.3; p trend = 0.10; TCN2 259 Arg/Arg variant MV OR = 3.82; 95% CI, 1.02–14.4; p trend = 0.06). The one-carbon metabolism genotypes were not significantly associated with LINE-1 methylation, although there were modest differences in mean LINE-1 methylation levels between certain genotypes. Collectively, these exploratory data provide suggestive evidence for the association of MTHFR 429 Ala/Ala and TCN2 259 Arg/Arg and CIMP status in colorectal cancer.
Similar content being viewed by others
Abbreviations
- BHMT:
-
Betaine–homocysteine methyltransferase
- BMI:
-
Body mass index
- CI:
-
Confidence interval
- CIMP:
-
CpG island methylator phenotype
- HPFS:
-
Health Professionals Follow-up Study
- LINE-1:
-
Long interspersed nucleotide element-1
- MSI:
-
Microsatellite instability
- MTHFR:
-
Methylenetetrahydrofolate reductase
- MTRR:
-
5-Methyltetrahydrofolate–homocysteine methyltransferase reductase (methionine synthase reductase)
- NHS:
-
Nurses’ Health Study
- nsSNP:
-
Nonsynonymous single nucleotide polymorphism
- OR:
-
Odds ratio
- SNP:
-
Single nucleotide polymorphism
- TCN2:
-
Transcobalamin 2
References
Esteller M (2008) Epigenetics in cancer. N Engl J Med 358(11):1148–1159
Feinberg AP (2008) Epigenetics at the epicenter of modern medicine. Jama 299(11):1345–1350
Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M et al (1997) DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res 379(1):33–41
Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW et al (2003) Induction of tumors in mice by genomic hypomethylation. Science 300(5618):489–492
Yamada Y, Jackson-Grusby L, Linhart H, Meissner A, Eden A, Lin H et al (2005) Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc Natl Acad Sci USA 102(38):13580–13585
Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C et al (2006) Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res 66(17):8462–9468
Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K et al (2006) Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 9(3):199–207
Karpf AR, Matsui S (2005) Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res 65(19):8635–8639
Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R et al (2007) LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS ONE 2(5):e399
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP (1999) CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 96(15):8681–8686
Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y et al (2007) Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA 104(47):18654–18659
Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ et al (2008) LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG methylator phenotype (CIMP) in colorectal cancer. Int J Cancer 122:2767–2773
Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD et al (2004) BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 53(8):1137–1144
Samowitz W, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA et al (2005) Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 129(3):837–845
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA et al (2006) CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 38(7):787–793
Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS et al (2007) IGFBP3 promoter methylation in colorectal cancer: relationship with microsatellite instability, CpG island methylator phenotype, and p53. Neoplasia 9(12):1091–1098
Slattery ML, Curtin K, Sweeney C, Levin TR, Potter J, Wolff RK et al (2007) Diet and lifestyle factor associations with CpG island methylator phenotype and BRAF mutations in colon cancer. Int J Cancer 120(3):656–663
Flood A, Caprario L, Chaterjee N, Lacey JV Jr, Schairer C, Schatzkin A (2002) Folate, methionine, alcohol, and colorectal cancer in a prospective study of women in the United States. Cancer Causes Control 13(6):551–561
Boyapati SM, Bostick RM, McGlynn KA, Fina MF, Roufail WM, Geisinger KR et al (2004) Folate intake, MTHFR C677T polymorphism, alcohol consumption, and risk for sporadic colorectal adenoma (United States). Cancer Causes Control 15(5):493–501
Giovannucci E (2004) Alcohol, one-carbon metabolism, and colorectal cancer: recent insights from molecular studies. J Nutr 134(9):2475S–2481S
Chen J, Kyte C, Valcin M, Chan W, Wetmur JG, Selhub J et al (2004) Polymorphisms in the one-carbon metabolic pathway, plasma folate levels and colorectal cancer in a prospective study. Int J Cancer 110(4):617–620
Murrell A, Heeson S, Cooper WN, Douglas E, Apostolidou S, Moore GE et al (2004) An association between variants in the IGF2 gene and Beckwith–Wiedemann syndrome: interaction between genotype and epigenotype. Hum Mol Genet 13(2):247–255
Curtin K, Slattery ML, Ulrich CM, Bigler J, Levin TR, Wolff RK et al (2007) Genetic polymorphisms in one-carbon metabolism: associations with CpG island methylator phenotype (CIMP) in colon cancer and the modifying effects of diet. Carcinogenesis 28(8):1672–1679
Koushik A, Kraft P, Fuchs CS, Hankinson SE, Willett WC, Giovannucci EL et al (2006) Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol Biomarkers Prev 15(12):2408–2417
Hazra A, Wu K, Kraft P, Fuchs CS, Giovannucci EL, Hunter DJ (2007) Twenty-four non-synonymous polymorphisms in the one-carbon metabolic pathway and risk of colorectal adenoma in the Nurses’ Health Study. Carcinogenesis 28(7):1510–1519
Colditz GA, Hankinson SE (2005) The Nurses’ Health Study: lifestyle and health among women. Nat Rev Cancer 5(5):388–396
Tranah GJ, Bugni J, Giovannucci E, Ma J, Fuchs C, Hines L et al (2006) O6-methylguanine-DNA methyltransferase Leu84Phe and Ile143Val polymorphisms and risk of colorectal cancer in the Nurses’ Health Study and Physicians’ Health Study (United States). Cancer Causes Control 17(5):721–731
Tranah GJ, Giovannucci E, Ma J, Fuchs C, Hunter DJ (2005) APC Asp1822Val and Gly2502Ser polymorphisms and risk of colorectal cancer and adenoma. Cancer Epidemiol Biomarkers Prev 14(4):863–870
Ogino S, Cantor M, Kawasaki T, Brahmandam M, Kirkner G, Weisenberger DJ et al (2006) CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 55:1000–1006
Livak KJ (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal 14(5–6):143–149
Devlin AM, Ling EH, Peerson JM, Fernando S, Clarke R, Smith AD et al (2000) Glutamate carboxypeptidase II: a polymorphism associated with lower levels of serum folate and hyperhomocysteinemia. Hum Mol Genet 9(19):2837–2844
Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D et al (2006) Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 8(2):209–217
Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS (2007) Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn 9(3):305–314
Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P et al (2008) Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res 68(20):8541–8546
Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP (2004) A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 32(3):e38
Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ et al (2008) LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer 122(12):2767–2773
Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D et al (2000) MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 28(8):E32
Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P (2006) Aberrant promoter methylation of p16 and MGMT genes in lung tumors from smoking and never-smoking lung cancer patients. Neoplasia 8(1):46–51
Alonso-Aperte E, Gonzalez MP, Poo-Prieto R, Varela-Moreiras G (2008) Folate status and S-adenosylmethionine/S-adenosylhomocysteine ratio in colorectal adenocarcinoma in humans. Eur J Clin Nutr 62(2):295–298
Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ et al (2002) A common mutation in the 5, 10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA 99(8):5606–5611
Giovannucci E, Stampfer MJ, Colditz GA, Rimm EB, Trichopoulos D, Rosner BA et al (1993) Folate, methionine, and alcohol intake and risk of colorectal adenoma. J Natl Cancer Inst 85(11):875–884
Sharp L, Little J, Brockton NT, Cotton SC, Masson LF, Haites NE et al (2008) Polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene, intakes of folate and related B vitamins and colorectal cancer: a case–control study in a population with relatively low folate intake. Br J Nutr 99(2):379–389
Brockton NT (2006) Localized depletion: the key to colorectal cancer risk mediated by MTHFR genotype and folate? Cancer Causes Control 17(8):1005–1016
Brockton NT (2008) Systemic folate status and risk of colorectal cancer. Cancer Causes Control 19(9):1005–1007 (author reply 3)
Weisberg IS, Jacques PF, Selhub J, Bostom AG, Chen Z, Curtis Ellison R et al (2001) The 1298A → C polymorphism in methylenetetrahydrofolate reductase (MTHFR): in vitro expression and association with homocysteine. Atherosclerosis 156(2):409–415
Kawakami K, Ruszkiewicz A, Bennett G, Moore J, Watanabe G, Iacopetta B (2003) The folate pool in colorectal cancers is associated with DNA hypermethylation and with a polymorphism in methylenetetrahydrofolate reductase. Clin Cancer Res 9(16 Pt 1):5860–5865
Oyama K, Kawakami K, Maeda K, Ishiguro K, Watanabe G (2004) The association between methylenetetrahydrofolate reductase polymorphism and promoter methylation in proximal colon cancer. Anticancer Res 24(2B):649–654
van den Donk M, van Engeland M, Pellis L, Witteman BJ, Kok FJ, Keijer J et al (2007) Dietary folate intake in combination with MTHFR C677T genotype and promoter methylation of tumor suppressor and DNA repair genes in sporadic colorectal adenomas. Cancer Epidemiol Biomarkers Prev 16(2):327–333
Yu J, Yu J, Almal AA, Dhanasekaran SM, Ghosh D, Worzel WP et al (2007) Feature selection and molecular classification of cancer using genetic programming. Neoplasia 9(4):292–303
Tsareva SA, Moriggl R, Corvinus FM, Wiederanders B, Schutz A, Kovacic B et al (2007) Signal transducer and activator of transcription 3 activation promotes invasive growth of colon carcinomas through matrix metalloproteinase induction. Neoplasia 9(4):279–291
Ateeq B, Unterberger A, Szyf M, Rabbani SA (2008) Pharmacological inhibition of DNA methylation induces proinvasive and prometastatic genes in vitro and in vivo. Neoplasia 10(3):266–278
Nagasaka T, Koi M, Kloor M, Gebert J, Vilkin A, Nishida N et al (2008) Mutations in both KRAS and BRAF may contribute to the methylator phenotype in colon cancer. Gastroenterology 134(7):1950–1960
Samowitz WS (2008) Genetic and epigenetic changes in colon cancer. Exp Mol Pathol 85(1):64–67
Sawan C, Vaissiere T, Murr R, Herceg Z (2008) Epigenetic drivers and genetic passengers on the road to cancer. Mutat Res 642(1–2):1–13
Thorstensen L, Lind GE, Lovig T, Diep CB, Meling GI, Rognum TO et al (2005) Genetic and epigenetic changes of components affecting the WNT pathway in colorectal carcinomas stratified by microsatellite instability. Neoplasia 7(2):99–108
Rocken C, Neumann K, Carl-McGrath S, Lage H, Ebert MP, Dierkes J et al (2007) The gene polymorphism of the angiotensin I-converting enzyme correlates with tumor size and patient survival in colorectal cancer patients. Neoplasia 9(9):716–722
Campos AC, Molognoni F, Melo FH, Galdieri LC, Carneiro CR, D’Almeida V et al (2007) Oxidative stress modulates DNA methylation during melanocyte anchorage blockade associated with malignant transformation. Neoplasia 9(12):1111–1121
Blanco D, Vicent S, Fraga MF, Fernandez-Garcia I, Freire J, Lujambio A et al (2007) Molecular analysis of a multistep lung cancer model induced by chronic inflammation reveals epigenetic regulation of p16 and activation of the DNA damage response pathway. Neoplasia 9(10):840–852
Kuester D, Dar AA, Moskaluk CC, Krueger S, Meyer F, Hartig R et al (2007) Early involvement of death-associated protein kinase promoter hypermethylation in the carcinogenesis of Barrett’s esophageal adenocarcinoma and its association with clinical progression. Neoplasia 9(3):236–245
Litkouhi B, Kwong J, Lo CM, Smedley JG III, McClane BA, Aponte M et al (2007) Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia 9(4):304–314
Lim U, Flood A, Choi SW, Albanes D, Cross AJ, Schatzkin A et al (2008) Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology 134(1):47–55
Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H et al (2008) Intra-individual change over time in DNA methylation with familial clustering. Jama 299(24):2877–2883
Hubner RA, Lubbe S, Chandler I, Houlston RS (2007) MTHFR C677T has differential influence on risk of MSI and MSS colorectal cancer. Hum Mol Genet 16(9):1072–1077
Anacleto C, Leopoldino AM, Rossi B, Soares FA, Lopes A, Rocha JC et al (2005) Colorectal cancer “methylator phenotype”: fact or artifact? Neoplasia 7(4):331–335
Acknowledgments
This research is supported by the National Institutes of Health Research Grants U54 CA100971, P01 CA87969, P01 CA55075, R01 CA070817, P50 CA127003, R03 CA142082, and K07 CA122826 (to S.O.); the Bennett Family Fund; and the Entertainment Industry Foundation’s National Colorectal Cancer Research Alliance. A.H. was supported in part by training grant NIH T-32 CA 09001-30. The content is solely the responsibility of the authors and does not necessarily represent the official views of NCI or NIH. Funding agencies did not have any role in the design of the study; the collection, analysis, or interpretation of the data; the decision to submit the manuscript for publication; or the writing of the manuscript. We deeply thank the Nurses’ Health Study and Health Professionals Follow-up Study cohort participants who generously agreed to provide us with biological specimens and information through responses to questionnaires; hospitals and pathology departments throughout the US for providing us with tumor tissue materials; Walter Willett, Sue Hankinson, and many other staff members who implemented and have maintained the cohort studies; Jean-Pierre Issa, Lanlan Shen, and Liying Yan for their assistance in the LINE-1 Pyrosequencing assay; and Peter Laird and Daniel Weisenberger for their assistance in the MethyLight assay.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hazra, A., Fuchs, C.S., Kawasaki, T. et al. Germline polymorphisms in the one-carbon metabolism pathway and DNA methylation in colorectal cancer. Cancer Causes Control 21, 331–345 (2010). https://doi.org/10.1007/s10552-009-9464-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10552-009-9464-2