Abstract
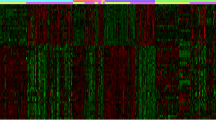
The aim of this study was systematic investigation of the differentially expressed genes during carcinogenesis in hepatocellular carcinoma (HCC) using cDNA microarray technology. The differentially expressed genes between 22 fresh HCC tissues and para-carcerous liver tissues (PCLT) were displayed using cDNA microarray technology. The result was verified by the reverse transcriptase polymerase chain reaction. Among the 8,464 human genes, 507 (5.99%) genes were expressed differentially at the mRNA levels between HCC and PCLT. Two hundred (2.36%) genes were down-regulated, whereas 307 (3.63%) genes were up-regulated. The mRNA expression levels of RhoC and protocadherin LKC detected by reverse transcription polymerase chain reaction (RT-PCR) were consistent with the microarray experiment. This study describes a gene expression profiling of HCC, which provides an extensive list of potential molecular markers for early diagnosis and molecular targets for the development of drugs to treat patients with primary HCC.


Similar content being viewed by others
References
Akriviadis EA, Llovet JM, Efremidis SC, et al. Hepatocellular carcinoma. Br J Surg. 1998;85:1319–1331. doi:10.1046/j.1365-2168.1998.00865.x.
Murray CJ, Lopez AD. Mortality by cause for eight regions of the world: global burden of disease study. Lancet. 1997;349:1269–1276. doi:10.1016/S0140-6736(96)07493-4.
Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43:S173–S181. doi:10.1002/hep.20956.
Huang G, Yang L, Yang J, Liu H, Yang Z. Correlation between epidermal growth factor and overexpression of vascular endothelial growth factor in hepatocellular carcinoma. Zhonghua Zhong Liu Za Zhi. 2002;24:564–566.
Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi:10.1126/science.286.5439.531.
Cherepinsky V, Feng J, Rejali M, Mishra B. Shrinkage-based similarity metric for cluster analysis of microarray data. Proc Natl Acad Sci USA. 2003;100:9668–9673. doi:10.1073/pnas.1633770100.
King HC, Sinha AA. Gene expression profile analysis by DNA microarrays: promise and pitfalls. JAMA. 2001;286:2280–2288. doi:10.1001/jama.286.18.2280.
Schena M. Genome analysis with gene expression microarrays. Bioessays. 1996;18:427–431. doi:10.1002/bies.950180513.
Li Y, Li Y, Tang R, et al. Discovery and analysis of hepatocellular carcinoma genes using cDNA microarrays. J Cancer Res Clin Oncol. 2002;128:369–379. doi:10.1007/s00432-002-0347-0.
Chen W, Fu X, Sun X, Sun T, Zhao Z, Sheng Z. Analysis of differentially expressed genes in keloids and normal skin with cDNA microarray. J Surg Res. 2003;113(2):208–216. doi:10.1016/S0022-4804(03)00188-4.
Okabe H, Satoh S, Furukawa Y, et al. Involvement of PEG10 in human hepatocellular carcinogenesis through interaction with SIAH1. Cancer Res. 2003;63:3043–3048.
Tsou AP, Chuang YC, Su JY, et al. Overexpression of a novel imprinted gene, PEG10, in human hepatocellular carcinoma and in regenerating mouse livers. J Biomed Sci. 2003;10:625–635.
Li CM, Margolin AA, Salas M, et al. PEG10 is a c-MYC target gene in cancer cells. Cancer Res. 2006;66:665–672. doi:10.1158/0008-5472.CAN-05-1553.
Ono R, Nakamura K, Inoue K, et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–106. doi:10.1038/ng1699.
Bafico A, Liu G, Goldin L, Harris V, Aaronson SA, et al. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi:10.1016/j.ccr.2004.09.032.
Patil MA, Chua MS, Pan KH, et al. An integrated data analysis approach to characterize genes highly expressed in hepatocellular carcinoma. Oncogene. 2005;24:3737–3747. doi:10.1038/sj.onc.1208479.
Ronski K, Sanders M, Burleson JA, Moyo V, Benn P, Fang M. Early growth response gene 1 (EGR1) is deleted in estrogen receptor-negative human breast carcinoma. Cancer. 2005;104:925–930. doi:10.1002/cncr.21262.
Clark EA, Golub TR, Lander ES, Hynes RO. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi:10.1038/35020106.
Suwa H, Ohshio G, Imamura T, et al. Overexpression of the RhoC gene correlates with progression of ductal adenocarcinoma of the pancreas. Br J Cancer. 1998;77:147–152.
Horiuchi A, Imai T, Wang C, et al. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab Invest. 2003;83:861–870.
Wang W, Yang LY, Yang ZL, Huang GW, Lu WQ. Expression and significance of RhoC gene in hepatocellular carcinoma. World J Gastroenterol. 2003;9:1950–1953.
Wang W, Yang LY, Huang GW, et al. Genomic analysis reveals RhoC as a potential marker in hepatocellular carcinoma with poor prognosis. Br J Cancer. 2004;90:2349–2355.
Okazaki N, Takahashi N, Kojima S, Masuho Y, Koga H. Protocadherin LKC, a new candidate for a tumor suppressor of colon and liver cancers, its association with contact inhibition of cell proliferation. Carcinogenesis. 2002;23:1139–1148. doi:10.1093/carcin/23.7.1139.
Acknowledgments
We are grateful to the following grants in support of this study: the National keystone basic research program of China (973 Program) (No. 2004CB720303), the Hi-Tech Research and Development Program of China (No.2006AA02Z4B2), the National Natural Science Foundation (No. 30801388), and the Distinguished Young Scholars of Xiangya Hospital CSU.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, W., Peng, J.X., Yang, J.Q. et al. Identification of Gene Expression Profiling in Hepatocellular Carcinoma Using cDNA Microarrays. Dig Dis Sci 54, 2729–2735 (2009). https://doi.org/10.1007/s10620-008-0667-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-008-0667-2