Abstract
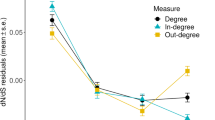
We examined the relationship between gene duplication, alternative splicing, and connectedness in a predicted genetic interaction network using published data from the nematode worm Caenorhabditis elegans. Similar to previous results from mammals, genes belonging to families with only one member (“singletons”) were significantly more likely to lack alternative splicing than were members of large multi-gene families. Genes belonging to multi-gene families lacking alternative splicing tended to have higher connectedness in the genetic interaction network than did genes in families that included one or more alternatively spliced members. Moreover, alternatively spliced genes were significantly more likely to interact with other alternatively spliced genes. These results support the hypothesis that certain key proteins with high degrees of network connectedness are subject to selection opposing the occurrence of alternatively spliced forms.



Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Ast G (2004) How did alternative splicing evolve? Nat Rev Genet 5:773–782
Barabási AL, Albert R (1999) Emergence of scaling in random networks. Science 286:509–512
Batagelj V, Mrvar A (2006) Pajek: program for analysis and visualization of large networks. Ljubljana
Blencowe BJ (2006) Alternative splicing: new insights from global analyses. Cell 126:37–47
Byrne AB, Weinrauch MT, Wong V, Koeva M, Dixon SJ, Stuart JM, Roy PJ (2007) A global analysis of genetic interactions in Caenorhabditis elegans. J Biol 6:8
Hollander M, Wolfe DA (1973) Nonparametric statistical methods. Wiley, New York
Hughes AL, Friedman R (2005) Gene duplication and the properties of biological networks. J Mol Evol 61:758–764
Kim E, Magen A, Ast G (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35:125–131
Kopelman NM, Lancet D, Yanai I (2005) Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet 37:588–589
Lareau LF, Green RE, Bhatnagar RS, Brenner SE (2004) The evolving roles of alternative splicing. Curr Opin Struct Biol 14:273–282
Papp B, Pál C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424:194–197
Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL (2002) Hierarchical organization of modularity in metabolic networks. Science 297:1551–1555
Rukov JL, Irimia M, Mørk S, Lund VK, Vinther J, Arctander P (2007) High qualitative and quantitative splicing in Caenorhabditis elegans and Caenorhabdidtis briggsiae. Mol Biol Evol 24:909–917
Stetefeld J, Ruegg MA (2005) Structural and functional diversity generated by alternative mRNA splicing. Trends Biochem Sci 30:515–521
Su Z, Wang J, Yu J, Huang X, Gu X (2006) Evolution of alternative splicing after gene duplication. Genome Res 16:182–189
Wagner A (2001) The yeast protein interaction network evolves rapidly and contains few redundant duplicated genes. Mol Biol Evol 18:1283–1292
Wagner A (2003) How the global structure of protein interaction networks evolves. Proc R Soc Lond B 270:457–460
Zhong W, Sternberg PW (2006) Genome-wide prediction of C. elegans genetic interactions. Science 311:1481–1484
Zhu X, Gerstein M, Snyder M (2007) Getting connected: analysis and principles of biological networks. Genes Dev 21:1010–1024
Acknowledgments
This research was supported by grant GM43940 from the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hughes, A.L., Friedman, R. Alternative splicing, gene duplication and connectivity in the genetic interaction network of the nematode worm Caenorhabditis elegans . Genetica 134, 181–186 (2008). https://doi.org/10.1007/s10709-007-9223-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-007-9223-9