Abstract
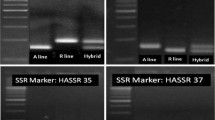
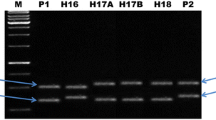
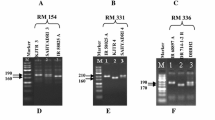
With an objective of achieving a breakthrough in the productivity of pigeonpea, a hybrid breeding technology based on elements of the cytoplasmic-nuclear male-sterility (CMS) system and partial natural out-crossing has recently been developed. However, there is no molecular diversity information available on parental lines of hybrids being generated at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT). This study deals with the use of 148 simple sequence repeat (SSR) markers, including 32 novel markers reported here for the first time, on 159 A (cytoplasmic male sterile), B (maintainer) and R (fertility restorer) lines. In total, 41 (27.7%) markers showed polymorphism with 2 to 6 (average 2.6) alleles and 0.01 to 0.81 (average 0.34) polymorphism information content (PIC) value. Of these polymorphic markers, 22 SSR markers showed polymorphism between A (ICPA 2039) and R (ICPR 2438) lines of the commercial hybrid (ICPH 2438); however, only 21 of these SSR markers showed the same profile between A (ICPA 2039) and B (ICPB 2039) lines. Finally, two SSR markers, CCB4 and CCttc006, were found most suitable for purity assessment of hybrid seeds of the ICPH 2438 hybrid. The utility of these two diagnostic SSR markers has been demonstrated by using seed lots of this hybrid from two sources, ICRISAT and Mahabeej. It is anticipated that molecular diversity information generated on parental lines of hybrids under development, and identification of the two most suitable markers for testing the purity of hybrid seeds of ICPH 2438, will facilitate the pigeonpea hybrid breeding programme.


Similar content being viewed by others
References
Ali MA, Seyal MT, Awan SI, Niaz S, Ali S, Abbas A (2008) Hybrid authentication in upland cotton through RAPD analysis. Aust J Crop Sci 2:141–149
Anderson JA, Churchill GA, Sutrique JE, Tanksley SD, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–186
Asif M, Rahman MU, Zafar Y (2006) Genotyping analysis of six maize (Zea mays L.) hybrid using DNA fingerprinting technology. Pakistan J Bot 38:1425–1430
Barbosa AMM, Geraldi IO, Benchimol LL, Garcia AAF, Souza CL Jr, Souza AP (2003) Relationship of intra- and inter-population tropical maize single cross hybrid performance and genetic distances computed from AFLP and SSR markers. Euphytica 130:87–99
Bohn M, Friedrich Utz H, Melchinger AE (1999) Genetic similarities among winter wheat cultivars determined on the basis of RFLPs, AFLPs and SSRs and their use for predicting progeny variance. Crop Sci 39:228–237
Burns MJ, Edwards KJ, Newbury HJ, Ford-Lloyd BV, Baggott CD (2001) Development of simple sequence repeat (SSR) markers for the assessment of gene flow and genetic diversity in pigeonpea (Cajanus cajan). Mol Ecol Notes 1:283–285
Chowdari KV, Venkatachalam SR, Davierwala AP, Gupta VS, Ranjekar PK, Govila OP (1998) Hybrid performance and genetic distance as revealed by the (GATA)4 microsatellite and RAPD markers in pearl millet. Theor Appl Genet 97:163–169
Cuc LM, Mace ES, Crouch JH, Quang VD, Long TD, Varshney RK (2008) Isolation and characterization of novel microsatellite markers and their application for diversity assessment in cultivated groundnut (Arachis hypogaea). BMC Plant Biol 8:55
Ferguson ME, Burow MD, Schultz SR, Bramel PJ, Paterson AH, Kresovich S, Mitchell S (2004) Microsatellite identification and characterization in peanut (A hypogaea L.). Theor Appl Genet 108:1064–1070
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Hashemi SH, Sayyed Mirmohammadi-Maibody AM, Nematzadeh GA, Arzani A (2009) Identification of rice hybrids using microsatellite and RAPD markers. Afr J Biotechnol 8:2094–2101
He G, Meng R, Newman M, Gao GM, Pittman RN, Prakash CS (2003) Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.). BMC Plant Biol 3:3
Jena KK, Pandey SK (1999) DNA markers for purification of A and B lines for hybrid rice improvement. Hybrid Rice News Lett 2:13–14
Jordan D, Tao Y, Godwin I, Henzell R, Cooper M, McIntyre C (2003) Prediction of hybrid performance in grain sorghum using RFLP markers. Theor Appl Genet 106:559–567
Kapila RK, Yadav RS, Plaha P, Rai KN, Yadav OP, Hash CT, Howarth CJ (2008) Genetic diversity among pearl millet maintainers using microsatellite markers. Plant Breed 127:33–37
Kidwell KK, Hartweck LM, Yandell BS, Crump PM, Brummer JE, Moutray J, Osborn TC (1999) Forage yields of alfalfa populations derived from parents selected on the basis of molecular marker diversity. Crop Sci 39:223–227
Kwon SJ, Ha WG, Wang HG, Yang SJ, Choi HC, Moon HP, Ahn SN (2002) Relationship between heterosis and genetic divergence in ‘Tongil’-type rice. Plant Breed 121:487–492
Lanza LLB, Souza CL Jr, Ottoboni LMM, Vieira MLC, Souza AP (1997) Genetic distance of inbred lines and prediction of maize single-cross performance using RAPD markers. Theor Appl Genet 94:1023–1030
Mishra GP, Singh RK, Mohapatra T, Singh AK, Prabhu KV, Zaman FU, Sharma RK (2003) Molecular mapping of gene for fertility restoration of wild abortive (WA) cytoplasmic male sterility using a basmati rice restorer line. J Plant Biochem Biotech 12:37–42
Moretzsohn MC, Leoi L, Proite K, Guimaraes PM, Leal-Bertioli SCM, Gimanes MA, Martin WS, Valls JFM, Grattapaglia D, Bertioli D (2005) A microsatellite-based, gene-rich linkage map for the AA genome of Arachis (Fabaceae). Theor Appl Genet 111:1060–1071
Nandakumar N, Singh AK, Sharma RK, Mohapatra T, Prabhu KV, Zaman FU (2004) Molecular fingerprinting of hybrids and assessment of genetic purity of hybrid seeds in rice using microsatellite markers. Euphytica 136:257–264
Naresh V, Yamini KN, Rajendrakumar P, Dinesh KV (2009) EST-SSR marker-based assay for the genetic purity assessment of safflower hybrids. Euphytica 170:347–353
Odeny DA, Jayashree B, Ferguson M, Hoisington D, Crouch J, Gebhardt C (2007) Development, characterization and utilization of microsatellite markers in pigeonpea. Plant Breed 126:130–137
Odeny DA, Jayashree B, Gebhardt C, Crouch J (2009) New microsatellite markers for pigeonpea (cajanus cajan (L.) millsp.). BMC Res Notes 2:35
Panguluri SK, Janaiah K, Govil JN, Kumar PA, Sharma PC (2006) AFLP fingerprinting in pigeonpea (Cajanus cajan (L.) Millsp.) and its wild relatives. Genet Resourc Crop Evol 53:523–531
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Enfield, Science Publishers, Montpellier, pp 43–76
Prasad M, Varshney RK, Roy JK, Balyan HS, Gupta PK (2000) The use of microsatellites for detecting DNA polymorphism, genotype identification and genetic diversity in wheat. Theor Appl Genet 100:584–592
Saxena KB (2008) Genetic improvement of pigeonpea—a review. Tropical Plant Biol 1:159–178
Saxena KB, Kumar RV, Srivastava N, Shiying B (2005) A cytoplsmic-nuclear male sterility system derived from a cross between Cajanus cajanifolius and Cajanus cajan. Euphytica 145:289–294
Saxena RK, Prathima C, Saxena KB, Hoisington DA, Singh NK, Varshney RK (2010a) Novel SSR markers for polymorphism detection in pigeonpea (Cajanus spp.). Plant Breed 129:142–148
Saxena RK, Saxena KB, Kumar RV, Hoisington DA, Varshney RK (2010b) SSR-based diversity in elite pigeonpea genotypes for developing mapping populations to map resistance to Fusarium wilt and sterility mosaic disease. Plant Breed 129:135–141
Smith JSC, Register JC III (1998) Genetic purity and testing technologies for seed quality: a company perspective. Seed Sci Res 8:285–293
Souframanien J, Manjaya JG, Krishna TG, Pawar SE (2003) Random amplified polymorphic DNA analyses of cytoplasmic male sterile and male fertile pigeonpea (Cajanus cajan (L.) Millsp.). Euphytica 129:293–299
Sundaram RM, Naveenkumar B, Biradar SK, Balachandran SM, Mishra B, IlyasAhmed M, Viraktamath BC, Ramesha MS, Sharma NP (2008) Identification of informative SSR markers capable of distinguishing hybrid rice parental lines and their utilization in seed purity assessment. Euphytica 163:215–224
Thudi M, Senthilvel S, Bottley A, Hash CT, Reddy AR, Feltus AF, Paterson AH, Hoisington DA, Varshney RK (2010) A comparative assessment of the utility of PCR-based marker systems in pearl millet. Euphytica. doi 10.1007/s10681-010-0148-5
Varshney RK, Penmetsa RV, Dutta S, Kulwal PL, Saxena RK, Datta S, Sharma TR, Rosen B, Carrasquilla-Garcia N, Farmer AD, Dubey A, Saxena KB, Gao J, Fakrudin B, Singh MN, Singh BP, Wanjari KB, Yuan M, Srivastava RK, Kilian A, Upadhyaya HD, Mallikarjuna N, Town CD, Bruening GE, He G, May GD, McCombie R, Jackson SA, Singh NK, Cook DR (2010) Pigeonpea genomics initiative (PGI): an international effort to improve crop productivity of pigeonpea (Cajanus cajan L.). Mol Breed. doi 10.1007/s11032-009-9327-2
Weber JL (1990) Informativeness of human (cC-dA)n (dG-dT)n polymorphisms. Genomics 7:524–530
Yang S, Pang W, Ash G, Harper J, Carling J, Wenzl P, Huttner E, Zong X, Kilian A (2006) Low level of genetic diversity in cultivated pigeonpea compared to its wild relatives is revealed by diversity arrays technology. Theor Appl Genet 113:585–595
Yashitola J, Thirumurugan T, Sundaram RM, Naseerullah MK, Ramesha MS, Sarma NP, Stone RV (2002) Assessment of purity of rice hybrids using microsatellite and STS markers. Crop Sci 42:1369–1373
Zhang Q, Gao YL, Yang SH, Ragab R, Saghai-Maroof MA, Li ZB (1994) A diallel analysis of heterosis in elite hybrid rice based on RFLPs and microsattelites. Theor Appl Genet 89:185–192
Acknowledgments
The study was funded by the Pigeonpea Genomics Initiative of the Indian Council of Agricultural Research (ICAR) under the framework of Indo-US Agricultural Knowledge Initiative (AKI), Generation Challenge Programme (www.generationcp.org) and Tropical Legume II (TL II) of Bill and Melinda Gates Foundation (BMGF).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saxena, R.K., Saxena, K. & Varshney, R.K. Application of SSR markers for molecular characterization of hybrid parents and purity assessment of ICPH 2438 hybrid of pigeonpea [Cajanus cajan (L.) Millspaugh]. Mol Breeding 26, 371–380 (2010). https://doi.org/10.1007/s11032-010-9459-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-010-9459-4