Abstract
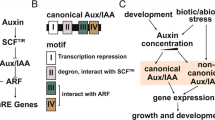
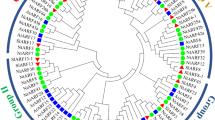
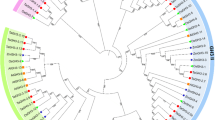
The phytohormone auxin is important in various aspects of organism growth and development. Aux/IAA genes encoding short-lived nuclear proteins are responsive primarily to auxin induction. Despite their physiological importance, systematic analysis of Aux/IAA genes in maize have not yet been reported. In this paper, we presented the isolation and characterization of maize Aux/IAA genes in whole-genome scale. A total of 31 maize Aux/IAA genes (ZmIAA1 to ZmIAA31) were identified. ZmIAA genes are distributed in all the maize chromosomes except chromosome 2. Aux/IAA genes expand in the maize genome partly due to tandem and segmental duplication events. Multiple alignment and motif display results revealed major maize Aux/IAA proteins share all the four conserved domains. Phylogenetic analysis indicated Aux/IAA family can be divided into seven subfamilies. Putative cis-acting regulatory DNA elements involved in auxin response, light signaling transduction and abiotic stress adaption were observed in the promoters of ZmIAA genes. Expression data mining suggested maize Aux/IAA genes have temporal and spatial expression pattern. Collectively, these results will provide molecular insights into the auxin metabolism, transport and signaling research.




Similar content being viewed by others
References
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95(5):707–735. doi:10.1093/aob/mci083
De Smet I, Jürgens G (2007) Patterning the axis in plants—auxin in control. Curr Opin Genet Dev 17(4):337–343. doi:10.1016/j.gde.2007.04.012
Kazan K, Manners JM (2009) Linking development to defense: auxin in plant–pathogen interactions. Trends Plant Sci 14(7):373–382. doi:10.1016/j.tplants.2009.04.005
Abel S, Theologis A (1996) Early genes and auxin action. Plant Physiol 111(1):9–17. doi:10.1104/pp.111.1.9
Walker JC, Key JL (1982) Isolation of cloned cDNAs to auxin-responsive poly (A)+ RNAs of elongating soybean hypocotyls. Proc Natl Acad Sci USA 79(23):7185–7189
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49(3–4):373–385
Tiwari SB, Hagen G, Guilfoyle TJ (2004) Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16(2):533–543. doi:10.1105/tpc.017384
Dreher KA, Brown J, Saw RE, Callisa J (2006) The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell 18(3):699–714. doi:10.1105/tpc.105.039172
Kepinski S, Leyser O (2004) Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci USA 101(33):12381–12386. doi:10.1073/pnas.0402868101
Worley CK, Zenser N, Ramos J et al (2000) Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J 21(6):553–562. doi:10.1046/j.1365-313x.2000.00703.x
Rogg LE, Lasswell J, Bartel B (2001) A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13(3):465–480. doi:10.1105/tpc.13.3.465
Reed JW (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci 6(9):420–425. doi:10.1016/S1360-1385(01)02042-8
Colón-Carmona A, Chen DL, Yeh KC, Abel S (2000) Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124(4):1728–1738. doi:10.1104/pp.124.4.1728
Song Y, You J, Xiong L (2009) Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol Biol 70(3):297–309. doi:10.1007/s11103-009-9474-1
Bennetzen JL, Chandler VL, Schnable PS (2001) National science foundation-sponsored workshop report. Maize genome sequencing project. Plant Physiol 127(4):1572–1578. doi:10.1104/pp.010817
Schnable PS, Ware D, Fulton RS et al (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326(5956):1112–1115. doi:10.1126/science.1178534
Finn RD, Tate J, Mistry J et al (2008) The Pfam protein families database. Nucleic Acids Res 36(Database issue):D281–D288. doi:10.1093/nar/gkm960
Eddy SR (2008) A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput Biol 4(5):e1000069. doi:10.1371/journal.pcbi.1000069
Letunic I, Copley RR, Schmidt S et al. (2004) SMART 4.0: towards genomic data integration. Nucleic Acids Res 32(Database issue):D142–D144. doi:10.1093/nar/gkh088
Hunter S, Apweiler R, Attwood TK et al. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37(Database issue):D211–D215. doi:10.1093/nar/gkn785
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Yi Chuan 29(8):1023–1026. doi:10.1360/yc-007-1023
Bailey TL, Boden M, Buske FA et al. (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37(Web Server issue):W202–W208. doi:10.1093/nar/gkp335
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. doi:10.1093/bioinformatics/btm404
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol 24(8):1596–1599. doi:10.1093/molbev/msm092
Ouyang Y, Chen J, Xie W, Wang L, Zhang Q (2009) Comprehensive sequence and expression profile analysis of Hsp20 gene family in rice. Plant Mol Biol 70(3):341–357. doi:10.1007/s11103-009-9477-y
Maher C, Stein L, Ware D (2006) Evolution of Arabidopsis microRNA families through duplication events. Genome Res 16(4):510–519. doi:10.1101/gr.4680506
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27(1):297–300. doi:10.1093/nar/27.1.297
Guilfoyle TJ, Hagen G (2007) Auxin response factors. Curr Opin Plant Biol 10(5):453–460. doi:10.1016/j.pbi.2007.08.014
Ding X, Hou X, Xie K, Xiong L (2009) Genome-wide identification of BURP domain-containing genes in rice reveals a gene family with diverse structures and responses to abiotic stresses. Planta 230(1):149–163. doi:10.1007/s00425-009-0929-z
Ma K, Xiao J, Li X, Zhang Q, Lian X (2009) Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene 444(1–2):33–45. doi:10.1016/j.gene.2009.05.018
Yue G, Hu X, He Y, Yang A, Zhang J (2010) Identification and characterization of two members of the FtsH gene family in maize (Zea mays L.). Mol Biol Rep 37(2):855–863. doi:10.1007/s11033-009-9691-3
Yang L, Zhu H, Guo W, Zhang T (2010) Molecular cloning and characterization of five genes encoding pentatricopeptide repeat proteins from Upland cotton (Gossypium hirsutum L.). Mol Biol Rep 37(2):801–808. doi:10.1007/s11033-009-9610-7
Xu P, Zhu HL, Xu HB et al (2010) Composition and phylogenetic analysis of wheat cryptochrome gene family. Mol Biol Rep 37(2):825–832. doi:10.1007/s11033-009-9628-x
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49(3–4):387–400
Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6(1):47–59. doi:10.1007/s10142-005-0005-0
Kalluri UC, Difazio SP, Brunner AM, Tuskan GA (2007) Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol 7:59. doi:10.1186/1471-2229-7-59
Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276(11):3148–3162. doi:10.1111/j.1742-4658.2009.07033.x
Acknowledgements
We are grateful to editors and reviewers for their helpful comments. We also thank Dr. Yidan Ouyang (Huazhong Agricultural University) for her help in HMM analysis. This work was supported partly by the Nature Science Foundation of Universities in Jiangsu Province (No. 09KJB180010) and the High-Level Personnel Foundation of Yangzhou University (No. nxy5286).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2010_58_MOESM3_ESM.tif
Multiple alignment of maize Aux/IAA proteins. Regions I, II, III and IV limited by black boxes represented four conserved domains of Aux/IAA proteins. Two NLSs and one βαα motif were indicated by bidirectional arrows. Phosphorylation sites were emphasized by filled triangles. Identical amino acids were highlighted by filled black columns. Positions of Aux/IAA proteins were showed by figures at both sides of sequences (TIFF 1116 kb)
11033_2010_58_MOESM4_ESM.tif
Phylogeny of Aux/IAA proteins. The phylogenetic tree indicated the relationship of maize, rice and Arabidopsis Aux/IAA proteins. Twelve paris of maize/rice Aux/IAA proteins were in the same clade of the phylogenetic tree. Contrarily, no maize/Arabidopsis Aux/IAA proteins were in the same clade of the dendrogram. Evolutional branches of maize/rice co-orthologs were marked in red in figure (TIFF 74 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Deng, D., Bian, Y. et al. Genome-wide analysis of primary auxin-responsive Aux/IAA gene family in maize (Zea mays. L.). Mol Biol Rep 37, 3991–4001 (2010). https://doi.org/10.1007/s11033-010-0058-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0058-6