Abstract
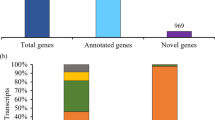
Transcriptome from high throughput sequencing-by-synthesis is a good resource of molecular markers. In this study, we present utility of massively parallel sequencing by synthesis for profiling the transcriptome of red pepper (Capsicum annuum L. TF68) using 454 GS-FLX pyrosequencing. Through the generation of approximately 30.63 megabases (Mb) of expressed sequence tag (EST) data with the average length of 375 base pairs (bp), 9,818 contigs and 23,712 singletons were obtained by raw reads assembly. Using BLAST alignment against NCBI non-redundant and a UniProt protein database, 30% of the tentative consensus sequences were assigned to specific function annotation, while 24% returned alignments of unknown function, leaving up to 46% with no alignment. Functional classification using FunCat revealed that sequences with putative known function were distributed cross 18 categories. All unigenes have an approximately equal distribution on chromosomes by aligning with tomato (Solanum lycopersicum) pseudomolecules. Furthermore, 1,536 high quality single nucleotide discrepancies were discovered using the Bukang mature fruit cDNA collection (dbEST ID: 23667) as a reference. Moreover, 758 simple sequence repeat (SSR) motif loci were mined from 614 contigs, from which 572 primer sets were designed. The SSR motifs corresponded to di- and tri- nucleotide motifs (27.03 and 61.92%, respectively). These molecular markers may be of great value for application in linkage mapping and association mapping research.






Similar content being viewed by others
References
Howard LR, Talcott ST, Brenes CH, Villalon B (2000) Changes in phytochemical and antioxidant activity of selected pepper cultivars (Capsicum species) as influenced by maturity. J Agr Food Chem 48(5):1713–1720. doi:10.1021/jf990916t
An CS, Kim SC, Go SL (1996) Analysis of red pepper (Capsicum annuum) genome. J Plant Biol 39:57–61
Huh JH, Kang BC, Nahm SH, Kim S, Ha KS, Lee MH, Kim BD (2001) A candidate gene approach identified phytoene synthase as the locus for mature fruit color in red pepper (Capsicum spp.). Theor Appl Genet 102(4):524–530. doi:10.1007/s001220051677
Rao GU, Ben Chaim A, Borovsky Y, Paran I (2003) Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor Appl Genet 106(8):1457–1466. doi:10.1007/s00122-003-1204-5
Lee CJ, Yoo E, Shin J, Lee J, Hwang HS, Kim BD (2005) Non-pungent Capsicum contains a deletion in the capsaicinoid synthetase gene, which allows early detection of pungency with SCAR markers. Mol Cells 19(2):262–267
Kim DS, Kim DH, Yoo JH, Kim BD (2006) Cleaved amplified polymorphic sequence and amplified fragment length polymorphism markers linked to the fertility restorer gene in chili pepper (Capsicum annuum L.). Mol Cells 21(1):135–140
Minamiyama Y, Tsuro M, Kubo T, Hirai M (2007) QTL analysis for resistance to Phytophthora capsici in pepper using a high density SSR-based map. Breed Sci 57(2):129–134. doi:10.1270/jsbbs.57.1291
Truong HTH, Kim KT, Kim S, Chae Y, Park JH, Oh DG, Cho MC (2010) Comparative mapping of consensus SSR markers in an intraspecific F8 recombinant inbred line population in Capsicum. Hort Environ Biotechnol 51(3):193–206
Schmid K, Törjék O, Meyer R, Schmuths H, Hoffmann M, Altmann T (2006) Evidence for a large-scale population structure of Arabidopsis thaliana from genome-wide single nucleotide polymorphism markers. Theor Appl Genet 112(6):1104–1114. doi:10.1007/s00122-006-0212-7
Lee GA, Koh HJ, Chung HK, Dixit A, Chung JW, Ma KH, Lee SY, Lee JR, Lee GS, Gwag JG, Kim TS, Park YJ (2009) Development of SNP-based CAPS and dCAPS markers in eight different genes involved in starch biosynthesis in rice. Mol Breeding 24(1):93–101. doi:10.1007/s11032-009-9278-7
Hu J, Nakatani M, Mizuno K, Fujimura T (2004) Development and characterization of microsatellite markers in sweetpotato. Breed Sci 54(2):177–188. doi:10.1270/jsbbs.54.177
Varshney RK (2009) Gene-based marker systems in plants: high throughput approaches for marker discovery and genotyping. In: Jain SM, Brar DS (eds) Molecular techniques in crop improvement. Springer, Netherlands, pp 119–142. doi:10.1007/978-90-481-2967-6_5
McCombie WR, Adams MD, Kelley JM, FitzGerald MG, Utterback TR, Khan M, Dubnick M, Kerlavage AR, Craig Venter J, Fields C (1992) Caenorhabditis elegans expressed sequence tags identify gene families and potential disease gene homologues. Nat Genet 1(2):124–131. doi:10.1038/ng0592-124
Ameline-Torregrosa C, Dumas B, Krajinski F, Esquerre-Tugaye MT, Jacquet C (2006) Transcriptomic approaches to unravel plant–pathogen interactions in legumes. Euphytica 147(1):25–36. doi:10.1007/s10681-006-6767-1
Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G, Ningre N, Renou J-P, Tamby JP, Le Thiec D, Hummel I (2010) Comparative transcriptomics of drought responses in populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11(1):630. doi:10.1186/1471-2164-11-630
Roberts GC, Smith CWJ (2002) Alternative splicing: combinatorial output from the genome. Curr Opin Chem Biol 6(3):375–383. doi:10.1016/s1367-5931(02)00320-4
Lee S, Yun SC (2006) The ozone stress transcriptome of pepper (Capsicum annuum L.). Mol Cells 21(2):197–205
Gore MA, Wright MH, Ersoz ES, Bouffard P, Szekeres ES, Jarvie TP, Hurwitz BL, Narechania A, Harkins TT, Grills GS, Ware DH, Buckler ES (2009) Large-scale discovery of gene-enriched SNPs. Plant Genome 2(2):121–133. doi:10.3835/plantgenome2009.01.0002
Barchi L, Bonnet J, Boudet C, Signoret P, Nagy I, Lanteri S, Palloix A, Lefebvre V (2007) A high-resolution, intraspecific linkage map of pepper (Capsicum annuum L.) and selection of reduced recombinant inbred line subsets for fast mapping. Genome 50(1):51–60. doi:10.1139/g06-140
Kim HJ, Nahm SH, Lee HR, Yoon GB, Kim KT, Kang BC, Choi D, Kweon O, Cho MC, Kwon JK, Han JH, Kim JH, Park MK, Ahn J, Choi S, Her N, Sung JH, Kim BD (2008) BAC-derived markers converted from RFLP linked to Phytophthora capsici resistance in pepper (Capsicum annuum L.). Theor Appl Genet 118(1):15–27. doi:10.1007/s00122-008-0873-5
Minamiyama Y, Tsuro M, Hirai M (2006) An SSR-based linkage map of Capsicum annuum. Mol Breeding 18(2):157–169. doi:10.1007/s11032-006-9024-3
Bonnet J, Danan S, Boudet C, Barchi L, Sage-Palloix AM, Caromel B, Palloix A, Lefebvre V (2007) Are the polygenic architectures of resistance to Phytophthora capsici and P. parasitica independent in pepper? Theor Appl Genet 115(2):253–264. doi:10.1007/s00122-007-0561-x
Ortega RG, Español CP, Zueco JC (1991) Genetics of resistance to Phytophthora capsici in the pepper line ‘SCM-334’. Plant Breeding 107(1):50–55. doi:10.1111/j.1439-0523.1991.tb00527.x
Chen YA, Lin CC, Wang CD, Wu HB, Hwang PI (2007) An optimized procedure greatly improves EST vector contamination removal. BMC Genomics 8(1):416. doi:10.1186/1471-2164-8-416
Li S, Chou HH (2004) Lucy2: an interactive DNA sequence quality trimming and vector removal tool. Bioinformatics 20(16):2865–2866. doi:10.1093/bioinformatics/bth302
Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucl Acids Res 32(18):5539–5545. doi:10.1093/nar/gkh894
Kim KY (2004) Developing one-step program (SSR MANAGER) for rapid identification of clones with SSRs and primer designing. MsD thesis. Seoul National University, Seoul
Thabuis A, Palloix A, Pflieger S, Daubèze AM, Caranta C, Lefebvre V (2003) Comparative mapping of Phytophthora resistance loci in pepper germplasm: evidence for conserved resistance loci across Solanaceae and for a large genetic diversity. Theor Appl Genet 106(8):1473–1485. doi:10.1007/s00122-003-1206-3
Weber APM, Weber KL, Carr K, Wilkerson C, Ohlrogge JB (2007) Sampling the Arabidopsis transcriptome with massively parallel pyrosequencing. Plant Physiol 144(1):32–42. doi:10.1104/pp.107.096677
Novaes E, Drost D, Farmerie W, Pappas G, Grattapaglia D, Sederoff R, Kirst M (2008) High-throughput gene and SNP discovery in Eucalyptus grandis, an uncharacterized genome. BMC Genomics 9(1):312. doi:10.1186/1471-2164-9-312
Livingstone KD, Lackney VK, Blauth JR, van Wijk R, Jahn MK (1999) Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152(3):1183–1202
Barbazuk WB, Scott JE, Hsin DC, Li L, Patrick SS (2007) SNP discovery via 454 transcriptome sequencing. Plant J 51(5):910–918. doi:10.1111/j.1365-313X.2007.03193.x
Hyten D, Cannon S, Song Q, Weeks N, Fickus E, Shoemaker R, Specht J, Farmer A, May G, Cregan P (2010) High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genomics 11(1):38. doi:10.1186/1471-2164-11-38
Parchman T, Geist K, Grahnen J, Benkman C, Buerkle C (2010) Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics 11(1):180. doi:10.1186/1471-2164-11-180
Lee HR, Bae IH, Park SW, Kim HJ, Min WK, Han JH, Kim KT, Kim BD (2009) Construction of an integrated pepper map using RFLP, SSR, CAPS, AFLP, WRKY, rRAMP, and BAC end sequences. Mol Cells 27(1):21–37. doi:10.1007/s10059-009-0002-6
Lee JM, Nahm SH, Kim YM, Kim BD (2004) Characterization and molecular genetic mapping of microsatellite loci in pepper. Theor Appl Genet 108(4):619–627. doi:10.1007/s00122-003-1467-x
Kim HJ, Baek KH, Lee SW, Kim JE, Lee BW, Cho HS, Kim WT, Choi D, Hur CG (2008) Pepper EST database: comprehensive in silico tool for analyzing the chili pepper (Capsicum annuum) transcriptome. Bmc Plant Biol 8(1):101. doi:10.1186/1471-2229-8-101
Yu JW, Dixit A, Ma KH, Chung JW, Park YJ (2009) A study on relative abundance, composition and length variation of microsatellites in 18 underutilized crop species. Genet Resour Crop Ev 56(2):237–246. doi:10.1007/s10722-008-9359-1
Yi G, Lee J, Lee S, Choi D, Kim BD (2006) Exploitation of pepper EST–SSRs and an SSR-based linkage map. Theor Appl Genet 114(1):113–130. doi:10.1007/s00122-006-0415-y
Acknowledgements
This study was supported by Agenda project (Grant No. 200901OFT071942004) of the Rural Development Administration (RDA), Republic of Korea. We are grateful for and acknowledge use of the tomato genome sequence, which was generated by the International Tomato Genome Sequencing Consortium (http://solgenomics.net/tomato/).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2011_1102_MOESM1_ESM.xls
Supplementary material 1: Annotation and FunCat results of all contigs and singletons in Capsicum annuum L. TF68 by BLAST against UniProt (XLS 11.632 kb)
11033_2011_1102_MOESM2_ESM.xls
Supplementary material 2: SNP discovery by mapping Capsicum annuum L. TF68 reads to NCBI Bukang cDNA library (XLS 892 kb)
11033_2011_1102_MOESM3_ESM.xls
Supplementary material 3: SSR discovery and primer information revealed by ARGOS program from Capsicum annuum L. TF68 contigs (XLS 219 kb)
Rights and permissions
About this article
Cite this article
Lu, FH., Cho, MC. & Park, YJ. Transcriptome profiling and molecular marker discovery in red pepper, Capsicum annuum L. TF68. Mol Biol Rep 39, 3327–3335 (2012). https://doi.org/10.1007/s11033-011-1102-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1102-x