Abstract
A fundamental problem in research on the origin of life is the process by which polymers capable of catalysis and replication were produced on the early Earth. Here we show that RNA-like polymers can be synthesized non-enzymatically from mononucleotides in lipid environments. The RNA-like polymers were initially identified by nanopore analysis, a technique with single molecule sensitivity. To our knowledge, this is the first such application of a nanopore instrument to detect RNA synthesis under simulated prebiotic conditions. The synthesis of the RNA-like polymers was confirmed by standard methods of enzymatic end labeling followed by gel electrophoresis. Chemical activation of the mononucleotides is not required. Instead, synthesis of phosphodiester bonds is driven by the chemical potential of fluctuating anhydrous and hydrated conditions, with heat providing activation energy during dehydration. In the final hydration step, the RNA-like polymer is encapsulated within lipid vesicles. This process provides a laboratory model of an early stage of evolution toward an RNA World.
Similar content being viewed by others
Introduction
Several studies have investigated possible pathways for the synthesis and degradation of RNA under hydrothermal conditions (White 1984; Larralde et al. 1995; Miller and Lazcano 1995; Kawamura et al. 1997; Levy and Miller 1998; Kawamura 2004). Kawamura’s results suggest that it is possible for synthesis of phosphodiester bonds to occur in hot aqueous solutions if chemically activated monomers and catalysts are present. We have previously shown that lipid vesicles can encapsulate oligomerization reactions (Chakrabarti et al. 1994) and can also provide an organizing template for the non-enzymatic polymerization of thioglutamic acid to peptides (Zepik et al. 2007). Here we report that the organizing effect of lipid systems can promote synthesis of RNA-like oligomers from non-activated 5′-nucleoside monophosphates.
Because polymerization by condensation is thermodynamically unfavorable in aqueous solutions, an energy source is required to drive phosphodiester bond formation. Imidazole esters of mononucleotides are commonly used as activated monomers and readily assemble on RNA templates to produce complementary RNA strands up to 30 nucleotides in length (Inoue and Orgel 1983; Orgel 1998). Huang and Ferris (2003) and Ferris (2002) found that the mineral surfaces of montmorillonite clay can organize chemically-activated mononucleotides so that RNA-like polymer chains in the 6–14 mer range are synthesized in the absence of templates, and up to 40–50 mers if a 10 mer is added as a primer or 1-methyladenine is used to activate the phosphate group of mononucleotides (Huang and Ferris 2006).
These conditions are useful models for investigating non-enzymatic polymerization mechanisms, but a plausible source of activated monomers in the prebiotic environment remains elusive. For this reason we are investigating other conditions that could drive polymer synthesis. We first note that phosphodiester bond formation is a relatively low-energy reaction. It was estimated that the standard free energy of synthesis is +5.3 kcal/mol (Dickson et al. 2000) which is similar to that of glucose-1-phosphate formation (+5.0 kcal/mol) from glucose and phosphate in solution. Thus, it should be possible to drive phosphodiester bond formation in the absence of activated substrates by producing conditions in which water can be removed from the reactants. More recently, Kawamura (2002) developed a method to monitor RNA synthesis and degradation of RNA under simulated hydrothermal vent conditions, and demonstrated that the rate of phosphodiester bond formation was faster than the rate of decomposition at 100°C, but at higher temperature ranges (200 and 300°C) degradation rates far exceeded synthesis. These results set an upper limit on thermal conditions for the origin of life, but also made it clear that there are no thermodynamic or kinetic barriers to RNA synthesis and stability in hyperthermophilic organisms like the chemolithoautotrophic archaeon Pyrolobus fumarii, which has been shown to be able to grow at 110°C (Stetter 1999). Other extremophiles have also been found to not only survive but thrive at such high temperatures (Stetter 1982; Kashefi and Lovely 2003).
In the present study we investigated RNA synthesis at elevated temperature ranges in fluctuating environments simulating hydrothermal springs that were likely to have been common in the prebiotic Earth. In such environments, cycles of wetting and drying in principle have sufficient free energy to “pump” a mixture of simple molecules toward increasingly complex molecular systems. The drying process concentrates otherwise dilute organic solutes, and also produces a chemical potential which can drive synthetic reactions such as ester bond formation. The reason this does not ordinarily work is that potential reactants are disorganized and immobilized within the solid matrix of a dry film, so that reactive groups only rarely come into contact to undergo condensation reactions. However, if a microenvironment could be discovered that not only organized the mononucleotides but also permitted diffusional mobility, it is possible that oligonucleotides resembling RNA would be synthesized from their monomers.
To this end, we are investigating the properties of microenvironments produced by organized fluid lipid matrices (liquid crystals). Ordering effects of amphiphilic structures in promoting prebiotic RNA synthesis have recently been discussed by Walde (2006). In past work, we have shown that solutes are trapped between the lamellae of lipid films when a mixture of lipid vesicles and a solute is dehydrated (Deamer and Barchfeld 1982). The vesicles fuse to form a multilamellar sandwich in which alternate lamellae contains a thin layer of the original solute. We are now testing the hypothesis that fluid lipid microenvironments impose order on mononucleotides in such a way that they are able to form extensive phosphodiester bonds and thereby produce RNA-like polymers sufficiently long to have catalytic activity (Szostak et al. 2001).
Methods
Materials
Mononucleotides (adenosine 5′-monophosphate and uridine 5′-monophosphate guanosine 5′-monophosphate and cytidine 5′-monophosphate), polyadenylic acid and polyuridylic acid were purchased from Sigma-Aldrich. The lipids, POPC (palmitoyl-oleoylphosphatidylcholine), POPA (palmitoyl-oleoylphosphatidic acid) and LPC (lysophosphatidylcholine) were purchased from Avanti Polar Lipids Inc. All other reagents were of analytical grade from Fisher, Sigma-Aldrich and Avanti Polar Lipids Inc.
Preparation of Lipid Dispersions
When phosphatidylcholine (POPC) was used, lipid was injected as a 20 mM ethanol solution into the aqueous phase to produce small unilamellar vesicles (Batzri and Korn 1973). Phosphatidic acid (POPA) and lysophosphatidylcholine (LPC enzymatically prepared from egg yolk phosphatidylcholine) were dispersed by 1 min agitation in a vortex stirrer. The lipid concentration was typically 10 mg/ml, and additions of AMP, UMP or 1:1 AMP:UMP mixtures (as 5′-mononucleoside phosphates) were adjusted to the desired mole ratio. For example, 10 mg/ml of POPC is 0.013 M and 10 mg/ml of AMP has a molarity of 0.027 M. In order to get a 1:2 molar ratio of POPC: AMP about equal volumes of 10 mg/ml stock solutions of each was added to the reaction setup.
Reaction Conditions
Mononucleotides and lipids (mole ratios of mononucleotide to lipid 2:1. 4:1 and 8:1) were mixed and put through a series of hydration-dehydration cycles to simulate a fluctuating environment on the prebiotic Earth. In a typical reaction the reactants were exposed to 1–7 cycles of wetting and drying in a volume of 0.5 ml. A stream of carbon dioxide (or in some experiments nitrogen gas) was used to dry the samples while they were exposed to varying experimental parameters including temperature (60–90°C), time (30–120 min), lipid composition and mole ratio of mononucleotide to lipid. After each drying cycle the samples were dispersed in 1 mM HCl and allowed to rehydrate for 15–20 min. During this time lipid vesicles reformed and components underwent mixing, and the mixtures were then exposed to a further dehydration cycle. The starting pH was 6.8, and this decreased to 2.2 at the end of seven cycles. When the cycle series was complete, the samples were dispersed in water, and the lipids were extracted twice with n-butanol (2:1 by volume) followed by hexane to remove excess remaining butanol. Some of the untreated samples were set aside for examination by light microscopy.
Gel Electrophoresis: 32P-labeling and Analysis of Reaction Products
After lipid extraction, samples were ethanol-precipitated and dissolved in 44 μl of water. For dephosphorylation, 1 μl of calf intestinal alkaline phosphatase (CIAP, 1 U/μl, MBI Fermentas) was added along with 5 μl of 10× CIAP buffer, and the reaction was incubated at 37°C for 30 min, followed by phenol extraction and ethanol precipitation. Glycogen (1 μl of stock 20 mg/ml) was added to facilitate precipitation of small amounts of RNA. The RNA aggregates were pelleted by centrifugation, then dissolved in 16 μl of water and labeled at the 5′-termini with 32P. Phosphorylation was carried out by adding 1 μl of T4 polynucleotide kinase (T4 PNK, 10 U/μl, New England Biolabs), 2 μl of 10× PNK buffer and 1 μl γ-[32P]ATP, followed by incubation at 37°C for 15 min. The end-labeled RNA-like polymers were purified by G50 spin columns (Amersham Biosciences) and stored at −20°C. For gel electrophoresis, 10 μl aliquots of the RNA samples were mixed with 3× denaturing loading solution (7 M urea, 10 mM EDTA, and 0.02% xylene cyanol and bromphenol blue) and separated by electrophoresis on 15% polyacrylamide gels containing 7 M urea, along with MW markers.
Reaction yields were determined by performing RiboGreen assays. The assay kit was obtained from BioTek Instruments, Inc., Winooski, Vermont. The RiboGreen RNA quantitation assay is a very sensitive technique that can detect as little as 1 ng/mL RNA. A standard curve was first obtained for polyadenyclic acid and was used to estimate the yields obtained in the experimental samples.
HPLC Analysis
HPLC analysis was performed by reverse phase chromatography employing an Alltima C-18 (5 μm) column (250 mm × 4.6 mm) on a Waters HPLC system. The samples were run at a flow rate of 0.5 ml/min in a water/acetonitrile gradient containing 0.1% trifluoroacetic acid (TFA). All the solvents used for this analysis were HPLC grade and obtained from Sigma-Aldrich (TFA) or Fischer Scientific (ACN).
Mass Spectrometry
Samples were purified by RP-HPLC and appropriate amounts were taken in 50% acetonitrile solution containing 1% formic acid. They were analyzed in the positive ion mode using electro spray ionization technique on a Micromass ZMD quadrupole mass analyzer. The samples were introduced via a Harvard apparatus Pump 11 at a flow rate of 30 μl/min. The solvents used were of HPLC grade obtained from Fischer Scientific.
Light Microscopy
Samples of the reaction mixture (15 μl) were stained with 0.5 mM ethidium bromide and 10 μl aliquots were examined by standard phase contrast and fluorescence microscopy methods at 400× magnification. To determine the extent to which hydrolysis of ester bonds in phospholipids could affect lipid bilayer stability, we also extracted phospholipids from the reaction mixture using equal volumes of 2:1 chloroform methanol. Aliquots of the chloroform phase (10 μl) were then dried on glass slides, rehydrated with 10 μl of water on a cover slip that then was allowed to settle over the dried lipid, and examined at 400× magnification by phase microscopy. This procedure was carried out after 1, 3 5 and 7 cycles.
Nanopore Analysis
A nanopore instrument was used for single molecule analysis of RNA samples. The detailed method is described in Akeson et al. (1999). Briefly, in the nanopore instrument, a U-shaped patch tube with a 30 μm diameter aperture is supported by a custom-made Teflon structure which contains two 90 μl wells connected through the patch tube. The wells were filled with 70 μl 1.0 M KCl-HEPES buffer, and a solution of diphytanoyl-sn-glycero-3-phosphocholine in hexadecane (25 mg/ml) was painted across the aperture to form a bilayer. Aliquots of alpha-hemolysin (10 μg/ml) were added to the cis side of the bilayer by pipetting and thoroughly mixed. Typically in 10–20 min a heptameric channel of hemolysin assembled and inserted into the bilayer, as detected by a steady 120 pA current at 120 mV applied potential. Samples to be analyzed were reconstituted in 14 μl of 1.0 M KCl/50 mM HEPES buffer, and applied to the cis side of the nanopore. When a linear polyanion such as single stranded RNA is captured by the electrical field in the pore, it is translocated through the pore by single molecule electrophoresis and its presence in the pore transiently blocks the ionic current. Translocation events were detected by an Axopatch 200B patch clamp amplifier and recorded using P-clamp 9.0 software. (Axon Instruments). Duration and amplitude were analyzed and plotted with Clampex and Clampfit software.
Results
Nanopore Analysis
We reasoned that yields would be low if RNA-like polymers long enough to have catalytic activity were synthesized from mononucleotides that were not chemically activated. We therefore used a nanopore instrument to analyze solutions in which polymerization may have taken place. Nanopore analysis can detect single linear polyanions such as RNA and DNA and therefore provides a highly sensitive method to scan solutions for products of a polymerization reaction (Kasianowicz et al. 1996; Akeson et al. 1999; Howorka et al. 2001; Meller and Branton 2002; Deamer and Branton 2002). As described in the “Methods” section, a commonly used nanopore is α-hemolysin, which self-assembles in a lipid bilayer into a heptamer containing a limiting aperture of 1.5 nm. The nucleic acid sample is then added to the cis compartment, and the electric field produced in the pore captures single-stranded nucleic acids which are translocated through the pore by electrophoresis. Each translocation event is detected as a characteristic blockade of the ionic current. The amplitude and duration of the blockade provide information about the composition and length of the nucleic acid strand. For instance, the length of polyadenylic acid homopolymers is directly related to the average duration of the polymer passage through the nanopore (Akeson et al. 1999), with typical translocation times of ~18 μs/base.
The first indication of RNA synthesis from AMP in a lipid environment was the appearance of ionic current blockades detected by the nanopore instrument. Figure 1 shows examples of blockades produced by known 50 mers of polyadenyclic acid compared with samples taken from a mixture of 5′-AMP and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) that had undergone 7 hydration-dehydration cycles. Previous investigations (Akeson et al. 1999) showed that individual polyadenylic acid molecules driven by electrophoresis through a nanopore blocked 85% of the ionic current through the pore. Translocation occurred at a rate of approximately 18–20 μs/base, so that the blockade signal of a 50 mer would typically be ~1 ms in duration. Examples of a typical blockade and a less common longer blockade are shown on the left in Fig. 1, and compared with several blockade signals from a sample of 5′-AMP subjected to 7 hydration-dehydration cycles in the presence of POPC. The blockades resembled those of polyadenylic acid 50 mers, but had a wider range of amplitude and duration due to the variable polymer length and chemical composition. In our experience such blockades can only be produced by linear polyanions such as single-stranded nucleic acids in the range of 20–100 mers. The fact that blockades were present was conclusive evidence that linear strands of an RNA-like polymer had been synthesized.
Nanopore analysis of RNA-like products. a blockades produced by a known 50 mer of polyadenylic acid. Typical blockades range around 1 ms in duration, representing a translocational velocity of 20 μs/base to pass through the nanopore. The presence of the RNA in the pore blocks approximately 85% of the ionic current that passes through the open channel. A few blockades last as much as ten times longer, but still have the same amplitude. b blockades produced by the RNA-like product from AMP:POPC (4:1) after five cycles. The duration and amplitude of the blockades had a greater range than the known 50 mer because of variations in chain length and conformation
Figure 2 shows examples of blockades produced by 5′-UMP under the same conditions, by 5′-AMP in the presence of phosphatidic acid (POPA) and by an equimolar mixture of all four 5′ nucleotides of RNA. These results indicate that the polymerization reaction is robust. Both purine and pyrimidine nucleotides can undergo polymerization, as can mixtures of all four nucleotides of RNA. Solutions of 5′-AMP alone (2.5 mM) did not produce blockades, nor did control experiments run in the absence of lipid (not shown).
Examples of blockades produced by 5′-UMP:POPC, 5′-AMP:POPA, and by equimolar mixtures of all four nucleotides in 4:1 ratios with POPA. UMP:POPC mixtures under the conditions of Fig. 1 tended to fall into two levels of blockade amplitude, and examples are shown here (top panel). Phosphatidic acid (POPA, center panel) was as effective as phosphatidylcholine (POPC) in promoting polymerization of 5’-AMP, and mixtures of all four nucleotides readily produced RNA-like molecules detected by their ionic current blockades (lower panel)
Although the presence of ionic current blockades provided qualitative evidence that RNA-like polymers were produced under the specified conditions in the presence of lipids, a more quantitative analytical approach is to plot blockade duration against amplitude of each blockades. Such plots are referred to as event diagrams, and provide statistical information about the results because hundreds of individual blockades can be compared as populations (Fig. 3). As noted earlier, no blockades are produced by AMP solutions by themselves (Fig. 3a). The few scattered events that are present near the origin are accounted for by electrical noise picked up by the instrument. Figure 3b shows the signals produced by 10 μM polyadenylic acid 50 mers. Note that the majority of events are present in a group with an average duration of 1 ms and 20 pA residual current, representing a blockade amplitude of 85%. This result is in accordance with previously reported data (Akeson et al. 1999). Figure 3c shows the event diagram for the RNA-like oligomers synthesized from 5′-AMP in the presence of POPA after 7 hydration–dehydration cycles. As would be expected, the blockades are considerably more variable in amplitude and duration than the 50 mer. This is due to the fact that the blockades are produced by oligomers having a wide range of chain lengths, as well as variable secondary structures produced by a random distribution of 2′–5′ and 3′–5′ phosphodiester bonds. Figure 3d–f show event diagrams for other combinations of nucleotides and lipids. It is interesting that the oligomers produced from UMP (Fig. 3e) separate into two groups in terms of blockade amplitude. This may be due to one species of oligomer only partially penetrating the nanopore, then falling out before full translocation has occurred.
Shows event diagrams in which blockade amplitude in picoamps is plotted against blockade duration in milliseconds. Each point represents the amplitude and duration of a single polymer molecule as it is translocated through the pore by an applied voltage of 120 mV. a Open channel current with no additions. A few short-lived low amplitude events seen in the control run are due to transient electronic noise. b Blockades produced by a known 50 mer of polyadenylic acid. The group of events between 0.1 and 1 ms represent complete translocations of RNA through the nanopore. Short duration and low amplitude events are due to the RNA entering the pore but then diffusing away before translocation occurs (Akeson et al. 1999). c and d Blockades produced by AMP:POPA and AMP:POPC reaction products had a range of amplitudes and durations. This is because the blockades are produced by mixed oligomers ranging from the minimal length detectable by nanopores (5–10 mers) to as long as 100 mers. Because each oligomer has variable numbers of 2′–5′ and 3′–5′ phosphodiester bonds, the durations and amplitudes of the blockades will be considerably more varied than those of the polyadenyclic acid 50 mer which has a specified length and only 3′–5′ bonds. e The event amplitudes produced by oligomers of 5′-UMP fall into two distinct groups having 25 and 50% blockades of ionic current. The reason is not yet understood. f A mixture of all four nucleotides reacting in the presence of POPA had a robust yield of products. Some of the blockade durations were very long, with a substantial number over 10 ms. This is probably due to secondary structures in the oligomers which must be unravelled during translocation and therefore increase the event duration
Yields of RNA-like Polymer
Because the nanopore results indicated that small amounts of polymers were synthesized from mononucleotides in the presence of lipids, we were interested in determining yields, length of the polymers, and the nature of the chemical bonds linking the monomers. Yields of the RNA-like polymers were determined by performing RiboGreen assays (BioTek, Inc., Winooski, Vermont). This method was chosen as a general quantitative approach for two reasons. First, it is highly sensitive and gives useful estimates of nanogram to microgram quantities of RNA such as polyadenylic acid. (The assay is much less sensitive to polyuridylic acid, so it was applied here only to products from AMP polymerization.) The second reason is related to the complexity of the polymeric products. From previous work on clay-catalyzed polymerization reactions, we expected that products were likely to be composed of RNA-like molecules having a broad range of chain lengths and variable amounts of 2′–5′ and 3′–5′ phosphodiester bonds within each molecule.
The total product ranged from 24 to 155 μg depending on several experimental variables, (Fig. 4) with the higher amount equivalent to ~6% yield of polymers by weight. Yields generally increased with the number of cycles the sample had experienced, typically reaching an apparent plateau after five cycles. Yields were highest when 1-palmitoyl-2-oleoylphosphatidic acid (POPA) was used (Fig. 2a), followed by lysophosphatidylcholine (LPC) and 1-palmitoyl-2-oleoylphosphatidylcholine (POPC). The yield obtained at 90°C was significantly greater than at 60 and 70°C (Fig. 2b). Base-catalyzed hydrolysis (0.1 M NaOH, 10 min, 60°) entirely hydrolyzed the RNA-like polymer to its component monomers as indicated by thin layer chromatography (not shown). This result excludes the possibility that bonds other than ester bonds were involved in the polymerization reaction.
Yields of RNA-like polymers obtained under different conditions. Experimental variables included the number of cycles and species of lipid (a), and temperature (b). In a, the monocleotide to lipid ratios were 2:1 (POPC), 1:1 (LPC) and 2:1 (POPA). In b the AMP to lipid ratio was 2:1. All reactions were carried out at 90°C
Although the yields are low when compared to those obtained with activated monomers, they are significant because the polymerization reaction can produce surprisingly long polymers using non-activated mononucleotides. The reason yields are low is most likely because energetically uphill phosphodiester bond formation during the anhydrous phase of the reaction cycle competes with hydrolysis during the hydrated phase. A similar competition between bond formation and hydrolysis would presumably be characteristic of reactions occurring in the prebiotic environment.
Gel Patterns of RNA-like Products
The results from nanopore analysis and RiboGreen assays were consistent with the possibility that linear strands of RNA-like polymers were synthesized in the presence of lipid. In order to confirm these observations and to determine the length of possible polymers, we used a procedure that produces radioactively labeled products for analysis by gel electrophoresis. The products were first treated with alkaline phosphatase to remove phosphate at the 5′ end, then labeled with γ-[32P]ATP using T4 polynucleotide kinase and analyzed by denaturing polyacrylamide gel electrophoresis.
This experiment was run multiple times using a variety of controls and conditions, including the number of cycles, species of lipid and nucleotide, nucleotide-to-lipid ratio and temperature. Figure 5a shows one such series in which the number of cycles was varied, using AMP and POPC in a 2:1 mole ratio. The amount of labelled RNA-like polymers increased steadily over seven cycles, which was consistent with the indications of the RiboGreen assay (Fig. 5a). Most of the RNA-like polymers ranged from 25 to 75 nucleotides in length, with a smaller fraction in the 100 mer range. This range of chain length was apparent even after a single cycle, and subsequent cycles served to increase the amount of polymer, but not the chain length. The labeled polymers shown in the gels consist of longer chains that were precipitated in ethanol, representing only a fraction of the total nucleotides initially present. The remainder, together with oligonucleotides shorter than 10 mers, was removed at this step in the procedure.
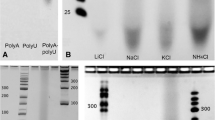
Gel patterns of RNA-like products end-labeled with AT32P. a shows the result of varying the number of cycles from 1 to 7. The reaction conditions were AMP:POPC 2:1. Several controls are also shown. Lane A: air was used instead of carbon dioxide during drying (7 cycles). Lane B: Lipid absent (7 cycles). Lane C: unheated control. Lane D: 10 µg of commercial polyadenylic acid as a positive control for the end labeling process. Lane E shows an RNA ladder containing known lengths of RNA in 10 nt increments. The effects of varying temperature (b), lipid (c), mononucleotide to lipid ratio (d), and mixtures of mononucleotides (e) were also investigated. (See text for details.) The abbreviations for lipid are POPC (1-palmitoyl-2-oleoylphosphatidylcholine) POPA (1-palmitoyl-2-oleoylphosphatidic acid) and LPC (egg lysophosphatidylcholine). The abbreviations for mononucleotide (AMP, 5′-adenosine monophosphate; UMP, 5′-uridine monophosphate) also indicate which lipid was used and the mole ratio of mononucleotide to lipid
A series of controls is shown in the lanes labeled A–D. If air was used instead of carbon dioxide in seven drying cycles, much less labeled polymer was observed (lane A). However, carbon dioxide and nitrogen atmospheres gave similar yields, so perhaps atmospheric oxygen in some way partially inhibits the reaction. If the experiment was run for seven cycles in the absence of lipid (lane B) or if cycling was not carried out (lane C) yields of labeled product were undetectable. Commercial polyadenylic acid was run as a positive control (lane D).
Figure 5b–f shows the effect of several experimental variables on the lipid-dependent RNA-like polymer synthesis reaction, which included temperature, substitution of different lipid and nucleotide species, and mixtures of nucleotides. Products were detected by end-labeling for all three temperature ranges tested (Fig. 5b) with the highest yields at 90°C. All three lipids promoted the condensation reaction (Fig. 5c) but products were much reduced in the absence of lipid. The nucleotide to lipid ratio affected both chain length and apparent yield (Fig. 5d). Substituting UMP for AMP seemed to have little effect on the yield of polymer (Fig. 5e) but the resulting chain lengths were markedly reduced in a 1:1 mixture of AMP and UMP, perhaps because the mixed purine and pyrimidine nucleotides are less stabilized by stacking energy than the pure nucleotides.
These results, taken together with the nanopore and electrophoresis results, confirmed that linear RNA-like polymers were the primary product of the reaction. Although we are confident that linear polymers are synthesized in the presence of lipid, it should be noted that the experimental conditions are much more complex than those of a typical chemical reaction. Each lane of reaction products shown in Fig. 5 represents a separate experiment, and during each wet–dry cycle there is a remixing of lipid, polymeric products and mononucleotide reactants. The reaction does not occur in solution, but instead in the interlamellar space of lipid lamellae in the dried film. This complexity could lead to considerable variation of yield from one sample to the next, and also to variation in the chemical nature of the phosphodiester bonds (both 2′–5′ and 3′–5′ bonds are present) and the 5′-ends of the RNA.
HPLC and Mass Spectrometry
HPLC and mass spectrometry are commonly used to analyze oligomeric RNA (Ferris 2002). However, for several reasons the oligomeric products reported here are less amenable to such analytical techniques. They have a range of lengths, as shown in the gel patterns above, and variable conformations due to random mixtures of 2′–5′ and 3′–5′ bonds. Furthermore, the only oligomers observed in gels were those with end groups that could be recognized by the two enzymes required for end labelling. Substantial amounts of oligomers with other end groups are likely to be present, such as cyclic phosphodiester bonds and even cyclic oligomers, rather than linear chains. We did carry out preliminary HPLC analysis, and could observe small amounts of products ranging up to 10 mers. Longer oligomers were either too low in concentration or too variable in composition to be separated and observed by HPLC as distinct peaks. Preliminary mass data indicated the presence of oligomers in the 20–30 mer range. A full analysis by HPLC and mass spectrometry will require large-scale preparations in order to yield oligomers in amounts sufficient to undergo further purification.
Microscopic Appearance and Lipid Stability
Membranous vesicles could be observed following rehydration after seven cycles of dehydration and heating at 90°C. A phase micrograph of such vesicles is shown in Fig. 6a. The fact that vesicles are visible demonstrates that fusion has occurred during the drying cycles, because the original vesicles were in the sub-micron size range and would not be resolved by phase microscopy. The same preparation was stained with 0.1 mM ethidium bromide, which intercalates into RNA structures and produces a fluorescent stain if RNA is present. Figure 6b shows a fluorescence image of the same sample. A diffuse fluorescence was pervasive, but some vesicles showed unstained interior volumes while others were brightly fluorescent. The presence of unstained vesicles lacking encapsulated material is predicted from the vesicle fusion that occurs during dehydration, because solutes are excluded from lamellar layers that were originally the interior of the lipid vesicles (Deamer and Barchfeld 1982). We cannot be certain of the nature of the vesicles with higher content of fluorescently stained substances, but if long strands of RNA-like molecules are in fact present, it is possible that some of the products may accumulate in aggregates, rather than being dispersed throughout the lipid phase. Similar aggregates could be seen in control preparations in which phosphatidylcholine vesicles were dried and rehydrated in the presence of biological RNA. Ethidium bromide staining produced no initial fluorescence in control lipid samples that were dried and then rehydrated in the absence of RNA (not shown).
We noticed in the micrographs that POPA vesicles, after several cycles, were somewhat disordered and aggregated, suggesting that partial degradation of the lipid was occurring. For this reason we followed the lipid composition by thin layer chromatography on silicic acid plates. The patterns clearly showed that after five cycles a significant fraction of POPA had hydrolyzed to lysophosphatidic acid and fatty acids, which appeared as separate spots on the plate. This was not the case with LPC and POPC vesicles which were significantly more stable to the wetting and drying cycles.
We also carried out microscopic examination of self-assembled lipid structures that form when dried lipid extracts are rehydrated (Figs. 7, 8 and 9). All of the lipids produced typical myelin figures and vesicles after one cycle of dehydration and heating, but after five cycles the hydrolysis occurring in phosphatidic acid samples was clearly affecting the self-assembly process. Bulk-phase crystals of fatty acid hydrolysis products (a mixture of palmitic and oleic acid) began to appear in the POPA samples following rehydration, while POPC and LPC were apparently unaffected. All three lipids were able to promote polymer synthesis (see Fig. 4a), so acid-catalyzed hydrolysis of the lipid does not seem to be a limiting factor in the reactions leading to polymer synthesis, at least after five cycles. The decreased yield observed after seven cycles in the presence of POPA (Fig. 4a) may be due to extensive hydrolysis of the phosphatidic acid to fatty acids. These do not form lamellar arrays of bimolecular membranes, and therefore are less able to promote the net synthesis of RNA-like polymers. Instead, the polymers present begin to undergo hydrolysis so that yields are reduced.
Effect of cycling on integrity of phosphatidylcholine. Lipid was extracted from samples that had undergone 1, 3, 5 and 7 cycles of dehydration – rehydration (a-d in the panel). An aliquot of the extract (10 µl) was dried on a microscope slide and allowed to self-assemble into membranous structures. It is clear that POPC maintained its ability to produce lipid bilayer structures, represented in the images as multilamellar tubular myelin figures and vesicles. Phase microscopy, 400× original magnification. Bar shows 20 µm
Effect of cycling on integrity of phosphatidic acid (POPA). In contrast to phosphatidylcholine, POPA began to undergo hydrolysis after 3 cycles (top right), and the reaction products – palmitic and oleic acid, and lysophosphatidic acid – began to form brushlike crystalline structures rather than membranes. These are clearly apparent after 5 and 7 cycles (lower panels). Phase microscopy, 400× original magnification. Bar shows 20 μm
Effect of cycling on integrity of lysophosphatidylcholine (LPC). This lipid, which has only one fatty acid on the 3-carbon, is relatively stable to hydrolysis. At low concentrations (<10 mM) LPC exists as micelles, but at the much higher concentrations of hydration from a dry phase it readily assembled into bilayer structures after one cycle (a) as well as after seven cycles (b) Phase microscopy, 400× original magnification. Bar shows 20 μm
Discussion
Fluctuating environments in the form of wet–dry cycles have long been considered as possible sources of free energy to drive uphill polymerization reactions (Kuhn 1976; Odom et al. 1979; Lahav et al. 1978; Lahav 1999). Verlander et al. (1973) showed that anhydrous heating of nucleotides could drive the formation of mixed 2′–5′ and 3′–5′ phosphodiester bonds, yielding dimers and trimers. Usher (1977) proposed that cycles of heating and drying, followed by rehydration, could drive phosphodiester bond formation and promote the accumulation of 3′–5′ bonds in the system due to the relative lability of 2′–5′ bonds to hydrolysis. The RNA-like polymers reported here differ from those of earlier studies in that the range of chain lengths is significantly longer (25–100 nucleotides) and the reaction does not require nucleotide activation to occur.
The reaction conditions in which RNA-like polymers form are relatively complex, and further research will be required before a mechanism can be put forward. It is likely that the process somehow involves an ordering effect of the lipid phase, presumably arising from the fact that the nucleotides are present at very high concentrations within lipid structures in the dry phase of each cycle. The combination of high concentration of reactants, interaction with polar head groups of lipids, and stacking of purine and pyrimidine bases would tend to align nucleotide molecules in such a way that phosphodiester bond formation is favored. Although the precise mechanism is not yet understood, one possibility is that at low pH ranges an –OH group on the phosphate becomes protonated to\( - {\text{OH}}_2^ + \) which then becomes a potential leaving group. A neighboring 2′ or 3′ hydroxyl of a ribose then can undergo a nucleophilic attack on the phosphorus to produce an ester bond. The moderately elevated temperature of 60–90°C provides activation energy for the reaction without significant degradation of reactants or products. Another possibility is that phosphodiester bond synthesis is driven by the initial formation of cyclic nucleotides which have sufficient stored energy in the internal diester bond to enter into polymerization reactions. These possibilities are now being investigated.
It may also be significant that, in contrast to the solid surface of a mineral such as clay, the lipid microenvironment is composed of fluid lipids. The diffusional mobility of mononucleotides adsorbed to a polar mineral surface such as clay would be markedly reduced, which would tend to slow reaction rates. However, reactant molecules captured within a fluid lipid matrix are able to diffuse and interact, which would promote covalent bond synthesis by condensation reactions. Another important difference between a multilamellar lipid phase and a solid bulk phase film of mononucleotides is that water molecules readily permeate lipid bilayers and are lost to the atmosphere, thereby reducing potential hydrolytic back reactions. In a solid bulk phase, water molecules cannot readily diffuse away from the reaction site.
The fact that lysophosphatidylcholine (LPC) also promoted polymerization of mononucleotides suggested an interesting possibility. In designing the experiments, we assumed that the nucleotide monomers would be present as 2-dimensional films between lipid bilayers. But LPC in dry or partially hydrated states forms a hexagonal I phase, in which the lipid molecules are arranged as cylinders around a central axis with head groups directed outward and tails inward (Reiss-Husson 1967). The term ‘hexagonal’ refers to the packing of the cylinders, which have an axis-to-axis spacing of 5 nm. In this structure, the mononucleotides would not occupy a 2-dimensional space, but instead would line up single file in the volume between the hexagonally packed cylinders. X-ray diffraction studies of lipid phases that have incorporated solutes during drying have not been carried out, particularly at elevated temperatures, but it seems possible that LPC and perhaps POPC and POPA exist in hexagonal phases when dried in the presence of solutes at elevated temperatures. This means that entrapped solutes such as AMP and UMP would be present in one-dimensional linear arrays, which would further order the molecular aggregates and thereby promote polymerization by phosphodiester bond formation.
It is perhaps surprising that polymeric products are able to survive the conditions of the cycling used here to drive polymer synthesis. For instance, at pH 3 and 100°C, conditions similar to those used in our experiments, 10% of biological RNA in solution is hydrolyzed in 40 min (Stanley 1968). In control experiments in the absence of lipid and mononucleotides, we found that polyadenylic acid is in fact hydrolyzed to monomers and oligomers after several cycles. However, polyadenylic acid in the presence of lipid was markedly less affected by these conditions, and after three cycles approximately 25% or the initial quantity remained. A certain amount of hydrolysis presumably does occur in the hydrated stage of a cycle, but when mononucleotides are present the forward reaction of phosphodiester bond synthesis would also occur in the anhydrous stage, with the net effect of preserving longer polymers. Kawamura and colleagues investigated the formation and stability of phosphodiesters bond at elevated temperatures (Kawamura et al. 1997; Kawamura 2004). Their findings show that net synthesis of oligonucleotides from activated mononucleotides can occur at 100°C because rates of formation of phosphodiester bonds at elevated temperatures exceed hydrolysis rates. This result is consistent with the observation reported here, that surprisingly long strands of RNA are maintained after synthesis at low pH and 90°C.
To summarize, lipid microenvironments are able to organize mononucleotides within a lipid matrix when phospholipid vesicles are mixed with mononucleotides and dried. Under these conditions, long strands of RNA-like molecules are synthesized by a condensation reaction when the reactants are exposed to one or more cycles of dehydration and elevated temperatures, followed by rehydration. The chemical potential driving the reaction is presumably supplied by the anhydrous conditions, with heat providing activation energy. At the end of the reaction, the polymers are encapsulated in vesicles formed by the lipid upon rehydration. It seems very plausible that fluctuating environments would be common on the prebiotic Earth, so that reactions such as those described here would produce membranous compartments containing RNA-like molecules if amphiphilic compounds and monomers were available. The encapsulated system of RNA-like polymers, in membranous compartments, could then undergo further chemical evolution toward the first forms of primitive cellular life.
References
Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW (1999) Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J 77:3227–3233
Batzri S, Korn ED (1973) Single bilayer liposomes prepared without sonication. Biochim Biophys Acta. 298:1015–1019
Chakrabarti AC, Breaker RR, Joyce GF, Deamer DW (1994) Production of RNA by a polymerase protein encapsulated within phospholipids vesicles. J Mol Evol 39:555–559
Deamer DW, Barchfeld GL (1982) Encapsulation of macromolecules by lipid vesicles under simulated prebiotic conditions. J Mol Evol 18:203–206
Deamer DW, Branton D (2002) Characterization of nucleic acids by nanopore analysis. Accounts Chem Res 35:817–825
Dickson KS, Burns CM, Richardson JP (2000) Determination of the free-energy change for repair of a DNA phosphodiester bond. J Biol Chem 275:15828–15831
Ferris J (2002) Montmorillonite catalysis of 30–50 mer oligonucleotides: laboratory demonstration of potential steps in the origin of the RNA world. Orig Life Evol Biosphere 32:311–332
Howorka S, Cheley S, Bayley H (2001) Sequence-specific detection of individual DNA strands using engineered nanopores. Nat Biotechnol 19:636–639
Huang W, Ferris JP (2003) Synthesis of 35–40 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA world. Chem Commun 21:1458–1461
Huang W, Ferris JP (2006) One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J Am Chem Soc 128:8914–8919
Inoue T, Orgel LE (1983) A nonenzymatic RNA polymerase model. Science 219:859–862
Kashefi K, Lovely DR (2003) Extending the upper temperature limit for life. Science 301:934
Kasianowicz J, Brandin E, Branton D, Deamer DW (1996) Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA 93:13770–13773
Kawamura K (2002) In situ UV–VIS detection of hydrothermal reactions using fused-silica capillary tubing within 0.08–3.2 s at high temperatures. Anal Sci 18:715–716
Kawamura K (2004) Behaviour of RNA under hydrothermal conditions and the origins of life. Intl J of Astrobiol 3:301–309
Kawamura K, Yosida A, Matumoto O (1997) Kinetic investigations for the hydrolysis of adenosine 5′-triphosphate at elevated temperatures: prospects for the chemical evolution of RNA. Viva Origino 25:177–190
Kuhn H (1976) Model consideration for the origin of life. Environmental structure as stimulus for the evolution of chemical systems. Naturwiss 63:68–80
Lahav N (1999) Biogenesis: theories of life’s origins. Oxford University Press, New York
Lahav N, White D, Chang S (1978) Peptide formation in the prebiotic era: thermal condensation of glycine in fluctuating clay environments. Science 201:67–69
Larralde R, Robertson MP, Miller SL (1995) Rates of decomposition of ribose and other sugars: implications for chemical evolution. Proc Natl Acad Sci USA 92:8158–8160
Levy M, Miller SL (1998) The stability of the RNA bases: implications for the origin of life. Proc Natl Acad Sci USA 95:7933–7938
Meller A, Branton D (2002) Single molecule measurements of DNA transport through a nanopore. Electrophoresis 23:2583–2591
Miller SL, Lazcano A (1995) The origin of life did it occur at high temperatures? J Mol Evol 41:689–692
Odom DG, Lahav N, Chang S (1979) Prebiotic nucleotide oligomerization in a fluctuating environment: effects of kaolinite and cyanamide. J Mol Evol 12:259–264
Orgel L (1998) Polymerization on the rocks: theoretical introduction. Orig Life Evol Biosphere 28:227–234
Reiss-Husson F (1967) Structure of liquid-crystalline phases of different phospholipids, monoglycerides, sphingolipids in the absence or presence of water. J Molec Biol 25:363–382
Stanley WM (1968) Fractionation of oligonucleotides according to degree of polymerization. Meth Enzymol 12:404–407
Stetter KO (1982) Ultrathin mycelia-forming organisms from submarine volcanic areas having an optimum growth temperature of 105°C. Nature 300:258–260
Stetter KO (1999) Extremophiles and their adaptation to hot environments. FEBS let 452:22–25
Szostak JW, Bartel DP, Luisi PL (2001) Synthesizing life. Nature 409:387–390
Usher DA (1977) Early chemical evolution of nucleic acids: a theoretical model. Science 196:311–313
Verlander MS, Lohrmann R, Orgel LE (1973) Catalysts for the self-polymerization of adenosine cyclic 2′, 3′-phosphate. J Mol Evol 2:303
Walde P (2006) Surfactant assemblies and their various possible roles for the origin(s) of Life. Orig Life Evol Biosph 109–150, Apr 27 (E-publication ahead of print)
White RH (1984) Hydrolytic stability of biomolecules at high temperatures and its implication for life at 250°C. Nature 310:430–432
Zepik HH, Rajamani S, Maurel M-C, Deamer DW (2007) Oligomerization of thioglutamic acid: encapsulated reactions and lipid catalysis. Orig Life Evol Biosph, Mar 25 (E-publication ahead of print)
Acknowledgements
The research reported here was supported by a grant from the NASA Exobiology program. We thank David Usher, Jack Szostak and Claude Bernasconi for helpful discussions during preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rajamani, S., Vlassov, A., Benner, S. et al. Lipid-assisted Synthesis of RNA-like Polymers from Mononucleotides. Orig Life Evol Biosph 38, 57–74 (2008). https://doi.org/10.1007/s11084-007-9113-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11084-007-9113-2