Abstract
Brachypodium distachyon (Brachypodium) has been recently recognized as an emerging model system for both comparative and functional genomics in grass species. In this study, 55,221 repeat masked Brachypodium BAC end sequences (BES) were used for comparative analysis against the 12 rice pseudomolecules. The analysis revealed that ~26.4% of BES have significant matches with the rice genome and 82.4% of the matches were homologous to known genes. Further analysis of paired-end BES and ~1.0 Mb sequences from nine selected BACs proved to be useful in revealing conserved regions and regions that have undergone considerable genomic changes. Differential gene amplification, insertions/deletions and inversions appeared to be the common evolutionary events that caused variations of microcolinearity at different orthologous genomic regions. It was found that ~17% of genes in the two genomes are not colinear in the orthologous regions. Analysis of BAC sequences also revealed higher gene density (~9 kb/gene) and lower repeat DNA content (~13.1%) in Brachypodium when compared to the orthologous rice regions, consistent with the smaller size of the Brachypodium genome. The 119 annotated Brachypodium genes were BLASTN compared against the wheat EST database and deletion bin mapped wheat ESTs. About 77% of the genes retrieved significant matches in the EST database, while 9.2% matched to the bin mapped ESTs. In some cases, genes in single Brachypodium BACs matched to multiple ESTs that were mapped to the same deletion bins, suggesting that the Brachypodium genome will be useful for ordering wheat ESTs within the deletion bins and developing specific markers at targeted regions in the wheat genome.
Similar content being viewed by others
Introduction
The large grass family (Poaceae), which includes major important cereals such as rice, wheat, and maize, encompasses over 10,000 species (Kellogg 2001). Early comparative genetic mapping based on RFLP markers has revealed considerable synteny between grass species, despite the great variation in genome size and evolutionary divergence times up to 60 million years (Moore et al. 1995; Gale and Devos 1998; Keller and Feuillet 2000). Because of the syntenic relationship among grass species, it is expected that the knowledge gained from an ideal model grass species will greatly facilitate the study of other important cereal crops. Rice was logically chosen as one model system for cereal crop genomics owing to its small genome size and importance as a staple crop (Goff 1999; Shimamoto and Kyozuka 2002; IRGSP 2005). The completion of the rice genome sequence has fueled its use in comparative genomics to understand the evolution of grass genomes as well as in map-based cloning of important genes in other cereal crops such as wheat (Song et al. 2002; Ling et al. 2003; Lai et al. 2004; Yan et al. 2004; Yan et al. 2006; Bruggmann et al. 2006; Wei et al. 2007).
Despite their close relationship, grass genomes are evolutionarily labile for many characteristics including chromosome number, ploidy, and genome size. In addition, common sequence changes such as insertion, deletion, duplication, and translocation can further complicate the use of many regions of the rice genome for cross-species comparison with other grasses (Sorrells et al. 2003; Bruggmann et al. 2006). Therefore, comparative analysis using more than two grass genomes could allow for elucidation of the nature of sequence changes that occurred in specific lineages during the evolutionary history of grass species (Song et al. 2002; Lai et al. 2004; Wei et al. 2007; Salse et al. 2008). For genome studies in Triticeae species including wheat and barley, a more closely related model grass genome will serve as a better intermediate for comparative and functional analysis.
The genus Brachypodium belongs to the Brachypodieae tribe, which is sister group to the four major cool season grass tribes of great economic importance—Triticeae, Aveneae, Poeae, and Bromeae (Draper et al. 2001; Kellogg 2001; Vogel et al. 2006). Hence, the Brachypodium genome is expected to show greater gene colinearity to the genomes of major cool season cereal grain and forage grasses and will be more useful in gene discovery in large Triticeae genomes such as wheat and barley (Garvin et al. 2008; Opanowicz et al. 2008; Ozdemir et al. 2008). In addition, the annual species in the genus Brachypodium possess a range of desirable biological features that make it well suited for functional genomics. For example, diploid Brachypodium has one of the smallest genomes in grasses (~300 Mb). The plant also has small physical stature, short life cycle, and undemanding growth requirements—all of which make it amenable to high-throughput genetic screening. Therefore, Brachypodium could serve as a utility model system for various types of plant research (Beckmann et al. 2008; Li et al. 2008; Idziak and Hasterok 2008; Parker et al. 2008; Spielmeyer et al. 2008).
Despite the growing interest in utilization of Brachypodium as a model grass, little is still known about the structure and organization of the Brachypodium genome. The utility of Brachypodium for grass crop genomics remains to be thoroughly tested. So far, comparative analysis with rice and wheat have been only conducted in a few genetic regions from a perennial, outbreeding species B. sylvativum (Foote et al. 2004; Griffiths et al. 2006; Bossolini et al. 2007; Faris et al. 2008; Spielmeyer et al. 2008), which has a genome reported to be ~470 Mb (Foote et al. 2004) with a size similar to that of rice, but larger than that of B. distachyon. B. sylvaticum has proved useful in mapping the wheat Ph1 gene (Griffiths et al. 2006). It was shown that the orthologous sequences from B. sylvaticum are more colinear to wheat as compared to those of rice. B. sylvaticum and wheat shared such high sequence identity that probes derived from B. sylvaticum sequences can be directly used for wheat mapping (Griffiths et al. 2006). Comparative studies on the wheat Lr34 region also indicated that B. sylvaticum and wheat showed perfect macro-colinearity of genetic markers, whereas rice contained a ~200-kb inversion in the orthologous region (Bossolini et al. 2007). It was estimated that B. sylvaticum and wheat diverged 35–40 Mya, significantly more recently than the divergence of rice and wheat (Bossolini et al. 2007). As far as we know, comparative analysis using B. distachyon genome has not been reported. A genome study on B. distachyon will provide important insights into the gene distribution and evolution of repetitive DNA in a compact grass genome.
Previously, we sequenced ~65,000 BAC ends from Brachypodium BAC libraries to generate ~38 Mb of random short genomic sequence, representing 10% of the Brachypodium genome (Huo et al. 2007). Analysis of BES revealed low repetitive DNA content and close phylogenetic relationship with the Triticeae species. In this study, we compared the BES against the rice genome to assess sequence conservation between these two compact grass genomes. To further analyze the colinearity of the orthologous regions, we sequenced nine Brachypodium BAC clones selected on basis of BAC-end matches to the rice genome. Our study provides the first comparison of the Brachypodium and rice genomes at multiple genetic loci. To evaluate the utility of Brachypodium for cereal crops with large genomes, the annotated Brachypodium genes were BLASTN searched against the wheat EST database and deletion-bin mapped wheat ESTs. Comparative analysis using the Brachypodium genome could offer a potentially useful strategy for the development of wheat genetic and linkage maps.
Materials and methods
Blast search of Brachypodium BES against the rice genome
To anchor Brachypodium BACs onto the rice genome, the 55,221 repeat masked BES were compared to the rice genome sequence using BLASTN. An expectation value (E) of e−10 was used as the significant threshold (www.tigr.org). The BES were assigned to individual rice chromosomes based on their best match to the rice genome. BES matching the rice genome were also analyzed by BLASTX against NCBI non-redundant protein database (http://www.ncbi.nlm.nih.gov/BLAST).
Sequencing of Brachypodium BAC clones and sequence assembling
BAC DNA was isolated with the Qiagen Large Construct Kit (Qiagen, Valencia, CA) and shotgun libraries were constructed as described previously (Gu et al. 2003). In brief, the purified DNA was sheared and size selected by agarose gel electrophoresis. Fragments with sizes between 3 and 5 kb were ligated into the pCR4TOPO vector using a TOPO cloning kit (Invitrogen, Carlsbad, CA). The ligation mixture was transformed into electrocompetent TOP10 cells. Plasmid DNA was isolated using the PerfectPrep Direct Bind Kit (Eppendorf, Boulder, CO). For each BAC, 768 subclones were sequenced from both ends using T7 and T3 primers and BigDye terminator chemistry (Applied Biosystems, Foster City, CA) on an ABI3730XL automated sequencer (Applied Biosystems). Gaps were filled by sequencing PCR products amplified directly from the BAC clones.
The BAC sequences were assembled using the Lasergene SeqMan Module (DNAStar, www.DNAStar.com) as described previously (Gu et al. 2003). In this module, we set the stringency for base calling and quality assessment to “high” to generate the most accurate consensus sequence possible. The sequence assembling was performed using a 40-bp window size and a 95% match requirement.
Annotation of Brachypodium repetitive elements
To define the Brachypodium repetitive element, a survey of the composition and contents of the Brachypodium repeat element sequences in the sequenced BACs was conducted using the RepeatMasker program (http://www.repeatmasker.org). The BAC sequences were also searched using BLASTN against the Triticeae Repeat Sequence (TREP) database (http://wheat.pw.usda.gov/ITMI/Repeats) and the local BLAST database containing unique Brachypodium repeat element sequences (Huo et al. 2007).
Gene annotation
To annotate coding sequences, a combination of BLASTN and BLASTX against non-redundant nucleotide and protein databases were utilized to identify all putative gene sequences. In addition, coding regions in the BAC sequences were also predicted using FGENESH (http://www.softberry.com) set for a monocot model. Predicted genes were then compared to the nonredundant and dbEST databases of NCBI (March 2008) using BLASTN and BLASTX. If a hypothetical protein was predicted, the sequence was searched against UniProt (Ver. 9.7) of European Bioinformatics Institute database (March 2008). Only matches with E values smaller than e−10 were accepted. The complete annotation of each sequenced BAC clone has been submitted to GenBank (Accession numbers EU730894-EU730902).
Brachypodium sequence comparison with rice and wheat
Orthologous rice sequences and annotations in VISTA format were downloaded from Gramene (http://www.gramene.org) and TIGR Rice Genome Browser (http://www.tigr.org). The orthologous rice sequences were re-annotated with the same criteria used to annotate the Brachypodium BACs. For comparative analysis between rice and Brachypodium, the rice CDS annotated in orthologous regions were aligned with the Brachypodium sequences to identify genes that were colinear. Sequence alignment analysis was performed using the VISTA program (Mayor et al. 2000). To compare Brachypodium and wheat, the annotated Brachypodium genes were compared to the deletion bin mapped wheat EST database (http://wheat.pw.usda.gov/wEST/) using BLASTN. Brachypodium genes were also compared to NCBI wheat and rice EST databases using BLASTN.
Results
Anchoring Brachypodium BES onto the rice genome
Analysis of ~65,000 Brachypodium BES revealed its relatively simple genome with low repetitive DNA content and high gene density (Huo et al. 2007). To provide an initial genome-wide comparison of rice and Brachypodium, both with compact genomes, we attempted to anchor Brachypodium BES onto the rice genome. Since the coding sequences of transposable elements (TE) often identify multiple noncolinear matches between the two grass species, it was critical to use repeat-masked BES in the analysis. Our BLASTN analysis showed that 14,547 out of 55,221 repeat-masked BES (26.3%) had matches to the rice genome sequence (E < e−10) at the nucleotide level. Further analysis using BLASTX showed that 11,982 (82.4%) out of these 14,547 BES matched to known protein-coding genes at E < e−10. The nature of the remaining 17.4% of the matches is unclear, however, some of these sequences could represent conserved noncoding sequences as previously suggested (Bossolini et al. 2007).
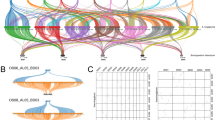
The matched BES were plotted along the 12 rice chromosomes to view the distribution pattern of these conserved sequences. The number of BES anchored to rice Chromosome 1 (chr1), chr2, and chr3 are higher than those aligning to the rest of the chromosomes. There were 2,018 (13.8%), 1,923 (13.2%) and 1,915 (13.2%) BES matched to the sequences on rice chromosome 1, 2 and 3, respectively, with a total of 40.2% of matched BES (Fig. 1a). More matches to these chromosomes are expected since they contain more genes than other chromosomes (Wu et al. 2002; IRGSP 2005). The number of matches to rice chr4 to chr12 ranged from 626 (4.3%) to 1247 (8.6%) (Fig. 1a).
Alignment of Brachypodium BES onto the rice genome. a Distribution of Brachypodium BES matched on 12 rice chromosomes. Repeat-masked Brachypodium BES was compared against the rice genome using BLASTN and BLASTX. The top match of each BES in BLASTN was used to determine its assignment to the rice chromosome. The number of total matches to each rice chromosome was counted and plotted. b Percentage of paired BES in each category. 1,734 BAC clones with paired ends both matching to the rice genome were categorized based on the distance of two matches on the same chromosome as indicated. BAC clones with two matches on different chromosomes are classified into category VI
Among the BES, there were 1,734 BAC clones for which the paired ends both showed significant matches to the rice genome. Since the approximate distance between these matches is known, these BAC clones are more informative in comparative studies to identify putative orthologous regions (Zhao et al. 2001, 2004). According to the positions of the two paired-end matches in the rice genome, these BACs can be placed into six different classes (Fig. 1b). For Classes I to V, both ends of the BAC clone matched rice sequences on the same chromosome. The distance separating the two ends ranged from less than 50 kb to over 1 Mb. For Class VI, the best matches of the two BAC ends to the rice sequences are on different chromosomes. The result showed that the number of BAC clones in each category was 116 (6.7%), 734 (42.3%), 58 (3.3%), 20 (1.2%), 153 (8.8%) and 653 (37.7%), respectively (Fig. 1b). We randomly selected 10 BAC clones from each category for determination of BAC insert size by CHEF gel electrophoresis. All 10 BAC clones in Class I contained an insert smaller than 50 kb, suggesting that the small distances between two matches in the rice genome reflected the small insert size in Brachypodium BAC clones. The BAC clones in other classes all had an insert with sizes ranging from 80 to 170 kb (data not shown), suggesting that most of the large size difference between a Brachypodium BAC and the orthologous rice region was the result of genomic changes that have occurred since the divergence of the two grass genomes.
BAC selection for sequencing
Considering the small insert size of Class I BACs, it is likely that most of the BACs in this category identified a colinear rice region with no major sequence rearrangements. This might be also true for many BACs in the Class II category, although some of the size difference between the Class II BAC insert and the corresponding rice region could be explained by the lower amount of repetitive DNA content in the smaller genome of Brachypodium (Huo et al. 2007). However, the large size differences observed in Class III, IV and V BACs and the scenario in Class VI BACs are likely due to substantial sequence difference between the two genomes. We examined the possibility that the two BAC ends in Class VI might match to the duplicated regions on two different rice chromosomes. 23.3% of these BACs showed that the two ends matched to two different regions originated from ancient genome duplication or chromosomal duplications in rice (Yu et al. 2005).
Dynamic sequence changes such as insertion, deletion, inversion, and translocation, coupled with rapid but differential repetitive DNA amplification and removal, are major evolutionary forces constantly reshaping individual plant genomes (SanMiguel et al. 1996; Vicient et al. 1999; Fu et al. 2002; Jiang et al. 2004; Kazazian 2004; Ma et al. 2004). In order to gain a more detailed view of the sequence changes in the regions represented by these paired BAC clones, we sequenced one or two BAC clones from different categories (Table 1). Considering the small insert size in Class I and likely colinearity with the corresponding rice region, no BAC clones were selected from this class. Instead, a BAC clone from Class II with an insert (~113 kb) similar to the size in the corresponding rice region (115.8 kb) was selected for sequencing. This BAC spanned a larger colinear region as compare to the BACs in Class I. Three random BACs, DH085B13, DB091J02 and DH002G02 were also chosen for sequencing in order to obtain a random sample sequence. The total assembled sequence of the nine Brachypodium BACs was 1,071 kb (Table 1).
Brachypodium repetitive DNA in sequenced BACs
Transposable elements (TE) are one of the major components of plant genomes (SanMiguel et al. 1996; Meyers et al. 2001; Li et al. 2004; Messing et al. 2004; Paux et al. 2006). To evaluate the TE content in the compact Brachypodium genome, the sequences from the nine BAC clones were compared to repetitive DNA databases using the RepeatMasker program (http://www.repeatmasker.org). TEs are classified based on their transposition mechanism as either DNA (Class II) or RNA (Class I) elements. The percentage of different types of TE is shown in Table 1. Clearly, there is significant variation of TE content and types among each BAC. The percentage of RNA TE ranges from 0.8% to 12.9% with an average of 6.7%. The percentage of DNA TE ranges from 0.9% to 3.0% with an average of 1.6%. Therefore, the average DNA TE content in the sequenced BACs is comparable to that estimated by BAC end sequences (1.28%); while the average RNA TE (6.7%) is slightly lower than the previous estimate (7.87%) (Huo et al. 2007).
Previously, we developed a database for the unique Brachypodium repetitive element sequences (UBRES) (Huo et al. 2007). The large contiguous BAC sequences were compared against the UBRES database (http://brachypodium.pw.usda.gov). Totally, 43,931 bp had significant matches to the UBRES. The percentage of the sequences matched to UBRES ranged from 1.0% to 9.8% with an average of 4.1%. This was lower than the percentage (7.4%) of UBRES observed previously (Huo et al. 2007).
Taken together, the total repetitive DNA content in sequenced BACs ranges from 4.2% to 23.5% with an average of 13.1%. This number is lower than the estimate based on BES (18.4%). This may be attributable to the pre-selection of six BAC clones that contained gene sequences at both ends. Relative higher gene content and lower DNA content in pre-selected BACs have been discussed (Devos et al. 2005). The difference may also be due to sampling error since the BAC sequences described here came from nine locations in the genomes whereas the BES were derived from the 32,500 locations sampled during our BES. Two of the random BACs have the highest repetitive DNA content. In addition, the large variation of repetitive DNA content in each BAC also indicates that TEs are not evenly distributed in the genome sequence. This suggests that there may be hot spots for TE insertions (Ma and Bennetzen 2004) even in the compact genome of Brachypodium.
Gene annotation of Brachypodium BACs and rice orthologous regions
The nine BAC clones represent different genetic loci in the Brachypodium genome. Detailed analysis of the sequences could provide the first insight into the gene content and distribution in the Brachypodium genome. As over-estimation of the gene number is a common problem in gene annotation (Bennetzen et al. 2004; Devos et al. 2005; Ma et al. 2005), we used stringent parameters for gene assignment in Brachypodium BACs. We only counted putative genes that had a significant match (BLASTX E value smaller than e−10) to a known gene that was not a transposable element (TE). For the nine Brachypodium BACs, a total of 196 genes were predicted by FGENESH, and 119 of these genes were confirmed by BLASTX search with these criteria. The gene annotations of each BAC are shown in Supplement 1.
The precise borders of each region in the rice genome orthologous to the individual Brachypodium BAC were identified by local BLAST of the BAC sequences against the rice genome assembly (TIGR rice pseudomolecule version 5). The detailed information of percentage of annotated repeat DNA and gene sequences in each BAC and its rice orthologous regions are shown in Fig. 2. According to both TIGR and Gramene databases, there were 214 genes annotated in these rice orthologous regions. When we used the same annotation strategy to re-annotate the orthologous rice regions, the gene number in the rice regions was decreased to 140 (Supplement 2). In general, a negative correlation of repetitive DNA content and gene number was observed in the Brachypodium BACs and the rice orthologous regions (Fig. 2).
Comparison of repeat DNA content and gene density in rice and Brachypodium. The repeat DNA content and the number of genes in sequenced Brachypodium BACs and corresponding orthologous regions of rice were analyzed. The sequence percentage taken by repeat DNA and genes was calculated for each orthologous region from rice and Brachypodium. The Brachypodium BAC clones indicated in x-axis was used to show the analysis results in the orthologous regions
The recent estimation of gene number in the rice genome is about 32,000 (Itoh et al. 2007), which is smaller than earlier estimates (Goff et al. 2002; Wu et al. 2002; Yu et al. 2005). It has been noted that many predicted hypothetical genes might be artificial (Itoh et al. 2007). Out of the originally annotated 214 genes, 75 either matched to the TIGR repeat database or had no match to the Arabidopsis protein database. These hypothetical genes were not present in the colinear regions in the Brachypodium BACs, providing further support that they were mis-annotated. Meanwhile, on rice chr6, a 3.9-kb non-annotated region (position 10008.6–10012.5 kb) was found to be similar to a gene encoding a serine/threonine kinase on Brachypodium BAC DB038O09. The sequence alignment using the VISTA program shows that they have 80% identity (data not shown). The TIGR rice transcript assembly TA64930_4530 supported our annotation; therefore a serine/threonine kinase gene was assigned to the rice region.
Gene density on selected Brachypodium BACs ranged from 6.8 kb to 16.7 kb/gene, with an average of 9.0 kb/gene (Table 1), whereas the gene density in the orthologous rice regions ranged from 6.2 kb up to 26.5 kb/gene with an average of 14.1 kb/gene. This number is much lower than the early estimation of gene density of 9.9 kb/gene in the rice genome (Wu et al. 2002; IRGSP 2005), and is close to the current estimation of 12.2 kb/gene (Itoh et al. 2007). However, if we consider the 75 hypothetical genes in rice, the average gene density in the rice regions would be 9.2 kb/gene, indicating that annotated gene density changes considerably depending on the gene annotation criterion used. As the same annotation criterion was applied to both Brachypodium BACs and the orthologous rice regions, our comparison of the gene density in these regions will provide a relatively unbiased result. In the rice regions orthologous to four Brachypodium BACs (DB009L22, DB038O09, DH003L20 and DB091J02), the difference in gene density is largely attributable to more repetitive DNA in the corresponding rice regions. However, in the other orthologous regions, the gene density is comparable (Fig. 2). These results suggest that the rice genome might have more regions containing a higher amount of repetitive DNA content. We found that large blocks of repetitive DNA regions were rarely found in Brachypodium sequences.
Comparison of orthologous regions of Brachypodium and rice
We compared the Brachypodium BAC sequences to the orthologous regions from rice. BAC DB009011 in Class II is 113-kb in length and contains 16 annotated genes. This region is orthologous to a 115.8-kb region on rice chr2 which contains 15 annotated genes. The VISTA alignments of the orthologous regions identified regions of sequence conservation and divergence (Fig. 3). Two Brachypodium genes, a vegetative storage (Gene2) and an ATP synthase (Gene15) gene, are absent in the rice region, while a rice gene (LOC_Os02g47850) is missing in the Brachypodium BAC. Thus, 14 genes are colinear with respect to order and orientation. Clearly, the most conserved regions are within the exon regions. The intergenic sequences are largely diverged although short regions of conserved non-coding sequences were detected in the colinear regions between Brachypodium and rice. The VISTA alignments easily showed the two non-shared genes (Gene2 and Gene15) that are only present in Brachypodium (Fig. 3). The intergenic distances are also similar in size to the orthologous region, suggesting a generally conserved region.
VISTA analysis of orthologous regions from Brachypodium and rice. The Brachypodium BAC DB009O11 sequence was used as backbone in comparison with the rice orthologous region. The rice orthologous sequence and annotation in VISTA format were downloaded from Gramene (http://www.gramene.org). Global alignment of the two sequences was performed using default settings. The x-axis represents the base sequence; the y-axis represents the percent identity of regions in two compared sequences. Genes annotated in Brachypodium BAC are only numbered sequentially in the figure. The annotation of each gene in Brachypodium and rice is provided in Supplement Table 1. Arrows indicate transcription orientation. Gene2 and Gene15 indicated in red in the Brachypodium BAC are nonshared in the rice orthologous region
The Class III BAC DB009L22 (128.8 kb) (Fig. 4b) and Class IV BAC DB038O09 (102.4 kb) (Fig. 4c) are orthologous to a 341.5-kb region on rice chr12 and 690-kb region on rice chr6, respectively. As shown in Fig. 4c, the large size difference (587.6 kb) between DB038O09 and the orthologous rice region is primarily due to the presence of a 426-kb fragment containing five genes in the rice genome. The absence of these five genes in the Brachypodium region also causes disruption of microcolinearity in the orthologous regions. Another factor causing the large size difference is the multiple duplications of a gene encoding UDP-glycosyltransferase, which resulted in nine copies (Gene16 to Gene24) spanning more than 100-kb in rice, whereas only one copy (Gene8) exists in the Brachypodium region (Fig. 4c). In addition to the above major sequence changes, there are three genes (Gene3, Gene14, and Gene15) unique to the rice region (Fig. 4c).
Comparison of the sequenced Brachypodium BAC regions with the rice orthologous regions. Sequenced Brachypodium BAC regions and the corresponding rice orthologous regions are represented by solid black lines. Genes conserved between Brachypodium and rice are depicted as blue boxes. Non-conserved genes are indicated in red and black boxes. Black boxes represent non-conserved genes at the end of BAC clones. Duplicated genes are labeled as green boxes. Orthologous genes in blue boxes are connected by dotted black lines. Annotated genes from both sequences were numbered sequentially. The annotation of each gene is provided in Supplement 1 and 2. For 4B and 4C, the yellow lines in the middle of the rice orthologous regions represented non-colinear fragments. In 4C, this region is re-drawn in scale on the top with annotated genes indicated. The pink box represents a gene that is missing in the rice genome annotation
The size difference between BAC DB009L22 and the orthologous region on rice chr12 was due to a 178-kb sequence containing three genes that was only present in the rice region (Fig. 4b). The presence of non-colinear 178-kb and 426-kb segments in the two orthologous rice regions could be caused by insertion in the rice genome or deletion in the Brachypodium genome.
Two BAC clones DH003L20 (89.3 kb) (Fig. 4d) and DB031O07 (107.3 kb) (Fig. 4e) were selected from the Class V category. The ends of DH003L20 matched two rice regions separated by 2,089 kb on chr6, while the ends of DB031O07 matched rice regions 7,032 kb apart on chr7. In both cases, it was found that the last genes on one end of the Brachypodium BAC clones were not present in the colinear positions in rice, instead they matched to regions millions of base pairs away on the same chromosome. The remaining regions in these two BACs were mostly colinear with the rice orthologous regions, except one Brachypodium gene in each BAC was not present in the orthologous rice regions (Supplement 1, 2).
The two ends of BAC DH037O21 (162.5 kb) (Fig. 4f) in Class VI matched to two regions located on different rice chromosomes. Further sequence analysis revealed that the region containing the first 11 genes in Brachypodium was orthologous to a 134.7-kb region on rice chr10, while the region containing the last 13 genes is orthologous to a 86.6-kb region on rice chr3 (Fig. 4f). Rice chr10 is colinear to rice chr3 due to ancient duplication events (Yu et al. 2005). Because of the differential sequence evolution in the duplicated regions, it appears that half of Brachypodium became more colinear with a region on rice chr3 and the other half more colinear with the paralogous region on rice chr10. If fact, we were unable to determine the true rice orthologous region for BAC DH037O21. Sequence changes have also occurred in the two rice regions as compared to the Brachypodium sequence. These include an inversion on chr3 and duplications of a proline-rich protein gene in two regions in rice as compared to one duplication region in Brachypodium (Fig. 4f). Furthermore, there are seven genes unique in the rice regions and three genes unique in the Brachypodium region.
Among the three random BACs, 20, 9, and 6 genes were identified in DH085B13 (138.9 kb) (Fig. 4g), DH002G02 (128.6 kb) (Fig. 4h), and DB091J02 (100.2 kb) (Fig. 4i), respectively. When compared with the rice orthologous regions, major sequence rearrangements were not identified, except a region containing three genes was inverted in Brachypodium BAC DH002G2. In addition, DH085B13 contains three non-colinear genes, DB091J02 and DH002G02 contain one non-colinear gene each (Supplement 1, 2).
Analysis of non-colinear genes
Our analysis showed that a total of 17 Brachypodium genes (out of 113 genes; 15%) are not present in the rice orthologous regions, while 27 rice genes (out of 140 genes; 19%) are absent in the Brachypodium regions. To examine if these non-colinear Brachypodium genes are present somewhere else in the rice genome, we BLASTN compared the non-colinear Brachypodium genes to the rice genome database. The result showed that 15 of the 17 non-colinear Brachypodium genes found matches in the rice genome at a BLASTX E value less than e−10, although it is not clear if these matches represent retrieving of the paralogous genes (Supplement 1, 2). Only two Brachypodium genes were missing from the rice genome; one gene is homologous to a wheat prolamine and the other is a gene fragment similar to wheat and Arabidopsis SKP1 gene. Among the 27 genes that were not shared in Brachypodium orthologous regions, 26 genes have significant matches in Brachypodium genome (e−10 or lower). Only one unknown gene LOC_Os03g26791 was missing in Brachypodium. Thus, our results are consistent with the previous results showing that most non-collinear genes in the maize or/and sorghum genomes were found in the rice genome at non-orthologous locations (Song et al. 2002; Lai et al. 2004). Nevertheless, based on the comparative sequence analysis, only ~17% of the genes in the two genomes are not colinear in the orthologous regions. It is worth noting that the degree of microsynteny based on sequence comparison in orthologous regions appeared to be different from that of macrosynteny estimated on BES alignment to the rice genome (Fig 1b). Although the reason causing the difference in microsynteny and macrosynteny levels is unclear, Gaut (2002) estimated that the macrosynteny probability of any given marker based on loss rate of syntenic gene during genome evolution is about 50% between two grass species with the divergence time of ~50 Mya, which is similar to the result observed in BES alignment to the rice genome. One possible explanation for the difference in our results could be attributable to the different analysis methods used. Detailed comparative sequence analysis in the orthologous regions allows identifying colinear genes that might have been dramatically changed due to high sequence divergence, rearrangement or partial deletion. In our BES alignment analysis, only the top match was counted. Therefore, the top match might be a paralogous gene on different chromosomes if the orthologous genes have been significantly changed, resulting in lower degree of synteny.
Sequence comparison among Brachypodium, rice and wheat
Several phylogenetic studies have indicated that Brachypodium and the Triticeae (wheat and barley) are more closely related to each other than to rice (Vogel et al. 2006; Bossolini et al. 2007; Huo et al. 2007). This close relationship suggests that Brachypodium may serve as a better model for Triticeae crop research. To evaluate the utility of Brachypodium as a model for wheat, we first compared all of our annotated Brachypodium genes to the wheat EST database. For the nine individual BACs, the percentages of genes with significant matches (E value less than e−5) ranged from 44.4% to 100%. Overall, out of the 119 annotated genes, 91 (76.5%) had significant matches to wheat ESTs (Table 2, Supplement 3). We also compared the annotated Brachypodium genes to the rice (Oryzeae) EST database. Only 63.9% of the genes matched to the rice EST database at E value less than e−5. Since only two Brachypodium genes were missing from the rice genome, the Brachypodium genes that were not present in the rice EST database must either be underrepresented in the rice tissues sampled for EST sequencing, or be pseudogenes. Since a similar number of wheat (1,051,465) and rice (1,220,261) ESTs were compared to the Brachypodium genes, most of the Brachypodium genes that did not match wheat EST are likely contained in the wheat genome.
To further evaluate the degree of divergence among Brachypodium, wheat, and rice, the number of synonymous substitutions (Ks) and nonsynonymous substitutions (Ka) per site between orthologous genes of Brachypodium, wheat, and rice were determined. To do so, only the coding sequences of the orthologous genes that can be identified for all three genomes were used in this analysis. The coding sequences for the orthologous wheat genes were obtained by BLASTN search against the Triticeae EST database. If the wheat and Brachypodium sequences showed more distantly related than Brachypodium and rice, the coding sequences was eliminated to avoid possibility that a paralogous wheat sequence or a pseudogene is used, which would distort the evolutionary distance (Bossolini et al. 2007). A total of eight genes met these criteria (Table 3). It appears that although the coding sequences of these genes were subjected to different rates of sequence evolution, the ratio of synonymous versus nonsynonymous substitution rates (Ka/Ks) for each gene was significantly less than 1 (P < 0.05; X-test of selection), suggesting they are all under purifying selection. Based on an average substitution rate of 6.5 × 10−9 mutations per synonymous site per year, as described for adh1 and adh2 coding region in grass (Gaut et al. 1996), the divergence time for Brachypodium and rice is ~49.3 Mya, it is ~37.8 Mya for Brachypodium and wheat, which is very similar to the estimation previously reported (Bossolini et al. 2007). This result supported the evidence that Brachypodium and wheat are more closely related than either to the rice.
The annotated Brachypodium genes were also compared to the deletion bin mapped wheat EST database (Qi et al. 2004). Eleven genes matched bin-mapped wheat ESTs (Table 3). If multiple genes from a single BAC matched the ESTs mapped to the same region, it could suggest that they identified the wheat orthologs. Three genes were from BAC DB009O11 (Class II), four from DH037O21 (Class VI), two from DB091J02 (random), and one each from DH002G02 (random) and DH085B13 (random). BAC DB009O11 had three genes that matched mapped wheat ESTs (BE497888, BE500611 and BE490512). These three ESTs have been mapped to the same co-localized bins (6AL4-0.55-0.90, 6BL5-0.40-1.00, and 6DL5-0.29-0.47) located on the long arm of wheat chromosome 6A, 6B and 6D, respectively (Qi et al. 2004; Randhawa et al. 2004). The order of bin-mapped ESTs is usually unknown. However, given the sequence conservation represented by BAC DB009O11 in Brachypodium and rice, the order of the three mapped wheat ESTs can now be tentatively assigned.
Four annotated genes (Gene5, Gene7, Gene12, and Gene19) from BAC DH037O21 matched to bin-mapped wheat ESTs (BI480570, BF482960, BE424589 and BG604404), respectively. Three of these ESTs (BI480570, BF482960, and BG604404) have map positions on wheat Chr4. The other EST BF424589 corresponding to Gene12 has been mapped to the short arm of Chr7 (7AS8-0.45-0.89, 7BS1-0.27-1.00). BAC DH037O21 represents a Class VI clone with its paired BES matched two rice regions from different chromosomes (Fig. 3b). However, considering that Gene5, Gene7, and Gene19 in the Brachypodium BAC DH037O21 are mapped to the same wheat chromosome (Chr4), but are located in different rice chromosomes (Fig. 4f), it is likely that Brachypodium and wheat will share more colinearity in this region.
The two wheat ESTs (BG274272 and BE517956) corresponding to the two annotated Brachypodium genes (Gene1 and Gene6) in BAC DB091J02, were mapped to deletion bins on different wheat chromosomes (C-4DL9-0.31 and C-5AS1-0.40). Clearly, colinearity is not retained based on these two mapped wheat ESTs. However, translocations involving wheat chromosomes 4A, 5A, and 7B, a paracentric inversion on chromosome 4A and a small pericentric inversion in centromeric bins on 5AS, 5BL and 5DL have been reported several times (Nelson et al. 1995; Linkiewicz et al. 2004; Qi et al. 2004). It was reported that part of the rice chromosome 3S is colinear with wheat 4BL/4DL, while the rest is colinear with wheat 5AL and 4AS (Buell et al. 2005). It seems that these regions were the conserved junctions that interrupt synteny blocks in each genome. The same conserved junction was reported in maize/sorghum/rice comparison (Bruggmann et al. 2006). Song and coworkers (2002) hypothesized that these regions were potential hotspots for chromosome changes. It is not clear what sequence changes were involved in BAC DB091J02 and the corresponding wheat region.
Discussion
The small size of the Brachypodium genome allowed us to use the high percentage of gene-containing BES for anchoring to a reference genome (rice). Using such a strategy has proved to be efficient in building whole-genome comparison (Larkin et al. 2003). Our results revealed that about 26.4% of repeat-masked BES matched to the rice genome and 82.4% of the matches (11,982 BES) were homologous to known genes. Comparative analysis using paired BAC ends help reveal local sequence changes in the orthologous regions. Our sequence analysis on six selected BACs and three random BACs provides the first snapshot view of the genome composition of Brachypodium and synteny conservation and divergence between Brachypodium, rice and wheat.
Composition and organization of Brachypodium genome
Rice and Brachypodium, diverged about 50 Mya, both have a compact genome and represent different lineages in the evolutionary path of grass species (Kellogg 2001). However, analysis of BES revealed that 12,113 BES had BLASTX matches to the non-redundant protein database at e−10 or smaller (Huo et al. 2007). Among them, 11,982 have significant matches (E < e−10) to the rice protein database, suggesting that over 82.6% of the protein-encoding genes are shared between rice and Brachypodium. A comparison of the rice and Arabidopsis protein sets revealed that 5,663 proteins are rice-specific and 3,402 Arabidopsis-specific (Itoh et al. 2007). The Brachypodium genome will allow us to determine which rice-specific genes are shared with Brachypodium and therefore, how many of these genes are monocot-specific genes.
Our analysis using paired BES was useful in identifying genomic regions that may have been subjected to considerable evolutionary changes. A sequence comparison of nine Brachypodium BACs to the orthologous rice regions identified specific sequence changes that have reshaped the orthologous regions of Brachypodium and rice. These sequence changes further validated the BES analysis results, suggesting that aligning the BES to a related, sequenced genome is an effective method to identify divergent regions. Comparative syntenic maps among grass species often only reveal large regions that are conserved on the basis of gene content and order with less emphasis on individual non-colinear genes. Our result showed that 15% of Brachypodium genes may fail to find their rice orthologs in the colinear rice regions and that 19% of rice genes are absent from the colinear positions in Brachypodium. Among the non-colinear genes, over 90% had at least one match in another part of the rice genome. However, it is difficult to determine if these genes moved to different locations or were lost in a genome-specific manner due to the presence of paralogous copies in the genome. In maize, at least 50% of the duplicated genes have been lost over a short period of time (Lai et al. 2004). Despite the considerable loss of duplicated genes, transposon-mediated gene movements have also been observed (Lal et al. 2003; Jiang et al. 2004; Lai et al. 2005), however, the importance of gene movements to the decay microcolinearity is not clearly understood.
The small genome of diploid Brachypodium is one of the important characteristics that make it an ideal model for large-genome grass species. The authoritative c value estimate of diploid Brachypodium is 0.36–0.39 (Bennett and Leitch 2005), and five different diploid accessions of Brachypodium have been identified with comparable c value (Vogel et al. 2006). Comparison to rice, with a c value of 0.51 pg (Bennett and Leitch 2005) and an accurate genome size of 389 Mb as determined by map-based sequencing (IRGSP 2005), yields an estimated genome size between 300 and 320 Mb for diploid Brachypodium. Our study further supports that the genome size of diploid Brachypodium is considerably smaller than that of rice. The repetitive DNA content observed in BES and the BAC sequences indicate that Brachypodium has less than 20% repetitive DNA, less than the rice genome (35%) (IRGSP 2005). The gene density observed in our Brachypodium BACs (~9.0 kb/gene) was higher than the gene density (~14 kb/gene) in the orthologous rice regions. Assuming that rice and Brachypodium have the same number of genes (estimated 32,000), the estimated genome size of Brachypodium would be slightly less than 300 Mb, much smaller than the recent estimation of 389 Mb for the rice genome (IRGSP 2005). A much better estimate of the Brachypodium genome size will be made after completion of the draft genome sequence (www.jgi.doe.gov). Genome size can vary considerably, even within the same genus. For example, two diploid Oryza species (Oryza sativa and Oryza australiensus) have genome size that differs by 2.7-fold (~390 Mb and ~975 Mb respectively). This larger size of Oryza australiensus genome is largely due to the rapid amplification of three LTR-retrotransposon families (Piegu et al. 2006). Thus, it is worth noting that Brachypodium sylvaticum, a perennial species, has an estimated genome size of 470 Mb (Foote et al. 2004), which is considerably larger than B. distachyon genome. Previous comparative sequence studies among wheat, rice, and Brachypodium were conducted using the Brachypodium sylvaticum sequence (Griffiths et al. 2006; Bossolini et al. 2007; Faris et al. 2008). In both Q gene and Lr34 resistance gene-containing regions, it was found that intergenic distances among colinear genes between B. sylvaticum and rice were generally larger in B. sylvaticum (Bossolini et al. 2007; Faris et al. 2008), suggesting it has a larger genome than that of rice. The result presented here indicates that B. distachyon has smaller intergenic regions and higher gene density than rice due largely to lower repetitive DNA content.
Comparative genomics improve genome annotation
Comparative genomics can complement other annotation methods (e.g. gene-finding program, BLAST search etc.) and help to provide a more accurate annotation (Katari et al. 2005). Comparative genomics aids discovery and annotation of gene structures and other functionally important sequences in both genomes. About 17% of predicted genes in both Arabidopsis (Katari et al. 2005) and rice (http://www.tigr.org/tdb/e2k1/osa1/riceInfo/info.shtml) were annotated hypothetical genes. Some of these hypothetical genes are artifacts of the annotation algorithms (Das et al. 1997). Therefore, validating hypothetical genes will greatly improve the precision of genome annotation. Katari and coworkers (2005) have confirmed 43 out of 110 Arabidopsis hypothetical proteins on the short arm of chromosome 4 by RT-PCR. They found that 46% of the hypothetical genes conserved in Brassica were expressed, whereas only 6% of the nonconserved hypothetical genes were expressed in Arabidopsis. They also pointed out that Brassica is more useful than rice in improving the annotation of the Arabidopsis genome because they are in the same family. The recent rice annotation project has identified that most previously annotated rice-specific proteins were hypothetical proteins (Itoh et al. 2007). The sequence of Brachypodium, along with the genome sequences from other grass species such as Sorghum, will help to verify these hypothetical genes in the rice genome. In this study, we identified 14 hypothetical proteins that were conserved in Brachypodium and rice. Among these fourteen genes, three (21.4%) had no homolog in Arabidopsis. On the other hand, Bossolini and coworkers (2007) have found that the percentage of the conserved genes increased when they re-annotated the rice region orthologous to wheat Lr34 locus region. They concluded that the apparent degree of conservation or colinearity of two compared genomes depends, in part, on the correct annotation of the compared sequence. In this study, 75 nonshared rice genes were removed as they matched TE or no Arabidopsis protein hit. We also observed the increasing percentage of the conserved genes between the two genomes.
Potential utility of Brachypodium for wheat genomics
Brachypodium has been proposed as a new model for the large-genome temperate grass crops because of its numerous desirable attributes, including a close relationship with Triticeae species. Several studies have shown that the relationship between Brachypodium and wheat is much closer than rice and wheat (Draper et al. 2001; Griffiths et al. 2006; Vogel et al. 2006; Bossolini et al. 2007; Huo et al. 2007). However, can Brachypodium really serve as a model for wheat? Bossolini et al. (2007) have doubted this because they found that only two-thirds of the genes from five wheat BACs on Lr34 locus were colinear with Brachypodium and relatively lower in gene density than that in the rice orthologous region. Conversely, Griffiths et al. (2006) in the course of mapping the wheat Ph1 candidate gene found that wheat and Brachypodium are more conserved, and that markers derived from Brachypodium sequences gave clear southern hybridization signals in wheat whereas markers made from rice sequence often failed.
We found that ~77% of Brachypodium genes have strong Triticeae EST matches (Table 2), and when matches were identified in wheat and rice EST databases, a higher matching score and lower E value to a wheat EST was usually found. These results suggest that Brachypodium sequences would be more useful for developing cross-species markers than the rice sequences. One potential strategy to improve wheat mapping is to identify wheat ESTs based on the annotation of colinear Brachypodium regions and to assess if they can be mapped onto the corresponding wheat genetic regions, thereby increasing the marker density. Furthermore, although 20% of the annotated Brachypodium genes have no matches in the Triticeae EST database, we can still confirm their genetic/physical locations in the wheat genome by directly using Brachypodium markers as has been demonstrated in the fine mapping of the complex Ph1 locus region in wheat (Griffiths et al. 2006).
The ideal model for wheat should share perfect microcolinearity with regard to gene content and order within a much compact genome. A few studies have shown the violation of microcolinearity between Brachypodium, wheat, and rice at local genomic regions. We can expect that the level of colinearity will not be homogenous along the chromosomes. For example, many resistance gene homologs are clustered in plant genome, and regions containing clusters of disease resistance sequences evolve more rapidly due to the frequent sequence exchanges than other regions containing house-keeping genes (Michelmore and Meyers 1998; Hulbert et al. 2001). Our result also indicated the translocation events specific in several wheat chromosome regions could have resulted in the disruption of colinearity between Brachypodium and wheat. The extent that Brachypodium can serve as a model species for genomics research on large genome grasses such as wheat is still unknown. Comparative analysis using the complete sequence of Brachypodium genome in the near future will provide an unprecedented view regarding the evolution of the grass genomes.
References
Beckmann M, Parker D, Enot DP, Duval E, Draper J (2008) High-throughput, nontargeted, metabolite fingerprinting using nominal mass flow injection electrophospray mass spectrometry. Nat Protocols 3:486–504. doi:10.1038/nprot.2007.500
Bennett MD, Leitch IJ (2005) Nuclear DNA amounts in angiosperms: progress, problems and prospects. Ann Bot (Lond) 95:45–90. doi:10.1093/aob/mci003
Bennetzen JL, Coleman C, Liu R, Ma J, Ramakrishna W (2004) Consistent over-estimation of gene number in complex plant genomes. Curr Opin Plant Biol 7:732–736. doi:10.1016/j.pbi.2004.09.003
Bossolini E, Wicker T, Knobel PA, Keller B (2007) Comparison of orthologous loci from small grass genomes Brachypodium and rice: implications for wheat genomics and grass genome annotation. Plant J 49:704–717. doi:10.1111/j.1365-313X.2006.02991.x
Bruggmann R, Bharti AK, Gundlach H, Lai J, Young S, Pontaroli AC, Wei F, Haberer G, Fuks G, Du C et al (2006) Uneven chromosome contraction and expansion in the maize genome. Genome Res 16:1241–1251. doi:10.1101/gr.5338906
Buell CR, Yuan Q, Ouyang S, Liu J, Zhu W, Wang A, Maiti R, Haas B, Wortman J, Pertea M et al (2005) Sequence, annotation, and analysis of synteny between rice chromosome 3 and diverged grass species. Genome Res 15:1284–1291. doi:10.1101/gr.3869505
Das S, Yu L, Gaitatzes C, Rogers R, Freeman J, Bienkowska J, Adams RM, Smith TF, Lindelien J (1997) Biology’s new Rosetta stone. Nature 385:29–30. doi:10.1038/385029a0
Devos KM, Ma J, Pontaroli AC, Pratt LH, Bennetzen JL (2005) Analysis and mapping of randomly chosen bacterial artificial chromosome clones from hexaploid bread wheat. Proc Natl Acad Sci USA 102:19243–19248. doi:10.1073/pnas.0509473102
Draper J, Mur LA, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge AP (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555. doi:10.1104/pp.010196
Faris JD, Zhang Z, Fellers JP, Gill BS (2008) Micro-colinearity between rice, Brachypodium, and Triticum monococcum at the wheat domestication locus Q. Funct Integr Genomics 8:149–164. doi:10.1007/s10142-008-0073-z
Foote TN, Griffiths S, Allouis S, Moore G (2004) Construction and analysis of a BAC library in the grass Brachypodium sylvaticum: its use as a tool to bridge the gap between rice and wheat in elucidating gene content. Funct Integr Genomics 4:26–33. doi:10.1007/s10142-003-0101-y
Fu H, Zheng Z, Dooner HK (2002) Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc Natl Acad Sci USA 99:1082–1087
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974. doi:10.1073/pnas.95.5.1971
Garvin DF, Gu YQ, Hasterok R, Hazen SP, Jenkins G, Mockler TC, Mur LAJ, Vogel JP (2008) Development of genetic and genomic research resources for Brachypodium distachyon, a new model system for grass crop research. Crop Sci 48:S69–S84. doi:10.2135/cropsci2007.06.0332tpg
Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154:15–28. doi:10.1046/j.1469-8137.2002.00352.x
Gaut BS, Morton BR, McCaig BC, Clegg MT (1996) Substitution rate comparisons between grasses and palms: synonymous rat different at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci USA 93:10274–10279. doi:10.1073/pnas.93.19.10274
Goff SA (1999) Rice as a model for cereal genomics. Curr Opin Plant Biol 2:86–89. doi:10.1016/S1369-5266(99)80018-1
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100. doi:10.1126/science.1068275
Griffiths S, Sharp R, Foote TN, Bertin I, Wanous M, Reader S, Colas I, Moore G (2006) Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439:749–752. doi:10.1038/nature04434
Gu YQ, Anderson OD, Londeore CF, Kong X, Chibbar RN, Lazo GR (2003) Structural organization of the barley D-hordein locus in comparison with its orthologous regions of wheat genomes. Genome 46:1084–1097. doi:10.1139/g03-071
Hulbert SH, Webb CA, Smith SM, Sun Q (2001) Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39:285–312. doi:10.1146/annurev.phyto.39.1.285
Huo N, Lazo GR, Vogel JP, You FM, Ma Y, Hayden DM, Coleman-Derr D, Hill TA, Dvorak J, Anderson OD, Gu YQ (2007) The nuclear genome of Brachypodium distachyon: analysis of BAC end sequences. Funct Integr Genomics 8:135–147
Idziak D, Hasterok P (2008) Cytogenetic evidence of nucleolar dominance in allotetraploid species of Brachypodium. Genome 51:387–391. doi:10.1139/G08-017
IRGSP (2005) The map-based sequence of the rice genome. Nature 436:793–800. doi:10.1038/nature03895
Itoh T, Tanaka T, Barrero RA, Yamasaki C, Fujii Y, Hilton PB, Antonio BA, Aono H, Apweiler R, Bruskiewich R et al (2007) Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res 17:175–183. doi:10.1101/gr.5509507
Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR (2004) Pack-MULE transposable elements mediate gene evolution in plants. Nature 431:569–573. doi:10.1038/nature02953
Katari MS, Balija V, Wilson RK, Martienssen RA, McCombie WR (2005) Comparing low coverage random shotgun sequence data from Brassica oleracea and Oryza sativa genome sequence for their ability to add to the annotation of Arabidopsis thaliana. Genome Res 15:496–504. doi:10.1101/gr.3239105
Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303:1626–1632. doi:10.1126/science.1089670
Keller B, Feuillet C (2000) Colinearity and gene density in grass genomes. Trends Plant Sci 5:246–251. doi:10.1016/S1360-1385(00)01629-0
Kellogg EA (2001) Evolutionary history of the grasses. Plant Physiol 125:1198–1205. doi:10.1104/pp.125.3.1198
Lai J, Ma J, Swigonova Z, Ramakrishna W, Linton E, Llaca V, Tanyolac B, Park YJ, Jeong OY, Bennetzen JL, Messing J (2004) Gene loss and movement in the maize genome. Genome Res 14:1924–1931. doi:10.1101/gr.2701104
Lai J, Li Y, Messing J, Dooner HK (2005) Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA 102:9068–9073. doi:10.1073/pnas.0502923102
Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC (2003) The maize genome contains a helitron insertion. Plant Cell 15:381–391. doi:10.1105/tpc.008375
Larkin DM, Everts-van der Wind A, Rebeiz M, Schweitzer PA, Bachman S, Green C, Wright CL, Campos EJ, Benson LD, Edwards J et al (2003) A cattle-human comparative map built with cattle BAC-ends and human genome sequence. Genome Res 13:1966–1972
Li W, Zhang P, Fellers JP, Friebe B, Gill BS (2004) Sequence composition, organization, and evolution of the core Triticeae genome. Plant J 40:500–511. doi:10.1111/j.1365-313X.2004.02228.x
Li X, Weng LK, Chapple C (2008) Improvement of biomass through lignin modification. Plant J 54:568–581. doi:10.1111/j.1365-313X.2008.03457.x
Ling HQ, Zhu Y, Keller B (2003) High-resolution mapping of the leaf rust disease resistance gene Lr1 in wheat and characterization of BAC clones from the Lr1 locus. Theor Appl Genet 106:875–882
Linkiewicz AM, Qi LL, Gill BS, Ratnasiri A, Echalier B, Chao S, Lazo GR, Hummel DD, Anderson OD, Akhunov ED et al (2004) A 2500-locus bin map of wheat homoeologous group 5 provides insights on gene distribution and colinearity with rice. Genetics 168:665–676. doi:10.1534/genetics.104.034835
Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci USA 101:12404–12410. doi:10.1073/pnas.0403715101
Ma J, Devos KM, Bennetzen JL (2004) Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res 14:860–869. doi:10.1101/gr.1466204
Ma J, SanMiguel P, Lai J, Messing J, Bennetzen JL (2005) DNA rearrangement in orthologous orp regions of the maize, rice and sorghum genomes. Genetics 170:1209–1220. doi:10.1534/genetics.105.040915
Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I (2000) VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16:1046–1047. doi:10.1093/bioinformatics/16.11.1046
Messing J, Bharti AK, Karlowski WM, Gundlach H, Kim HR, Yu Y, Wei F, Fuks G, Soderlund CA, Mayer KF, Wing RA (2004) Sequence composition and genome organization of maize. Proc Natl Acad Sci USA 101:14349–14354. doi:10.1073/pnas.0406163101
Meyers BC, Tingey SV, Morgante M (2001) Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res 11:1660–1676. doi:10.1101/gr.188201
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution. Grasses, line up and form a circle. Curr Biol 5:737–739. doi:10.1016/S0960-9822(95)00148-5
Nelson JC, Sorrells ME, Van Deynze AE, Lu YH, Atkinson M, Bernard M, Leroy P, Faris JD, Anderson JA (1995) Molecular mapping of wheat: major genes and rearrangements in homoeologous groups 4, 5, and 7. Genetics 141:721–731
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177. doi:10.1016/j.tplants.2008.01.007
Ozdemir BS, Hernandez P, Filiz E, Budak H (2008) Brachypodium genomics. Int J Plant Genomics 2008:536104
Parker D, Beckmann M, Enot DP, Overy DP, Rios ZC, Gilbert M, Talbot N, Draper J (2008) Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat Protocols 3:435–445. doi:10.1038/nprot.2007.499
Paux E, Roger D, Badaeva E, Gay G, Bernard M, Sourdille P, Feuillet C (2006) Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J 48:463–474. doi:10.1111/j.1365-313X.2006.02891.x
Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, Kim H, Collura K, Brar DS, Jackson S, Wing RA, Panaud O (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16:1262–1269. doi:10.1101/gr.5290206
Qi LL, Echalier B, Chao S, Lazo GR, Butler GE, Anderson OD, Akhunov ED, Dvorak J, Linkiewicz AM, Ratnasiri A et al (2004) A chromosome bin map of 16, 000 expressed sequence tag loci and distribution of genes among the three genomes of polyploid wheat. Genetics 168:701–712. doi:10.1534/genetics.104.034868
Randhawa HS, Dilbirligi M, Sidhu D, Erayman M, Sandhu D, Bondareva S, Chao S, Lazo GR, Anderson OD, Miftahudin et al (2004) Deletion mapping of homoeologous group 6-specific wheat expressed sequence tags. Genetics 168:677–686. doi:10.1534/genetics.104.034843
Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C (2008) Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell 20:11–24. doi:10.1105/tpc.107.056309
SanMiguel P, Tikhonov A, Jin YK, Motchoulskaia N, Zakharov D, Melake-Berhan A, Springer PS, Edwards KJ, Lee M, Avramova Z, Bennetzen JL (1996) Nested retrotransposons in the intergenic regions of the maize genome. Science 274:765–768. doi:10.1126/science.274.5288.765
Shimamoto K, Kyozuka J (2002) Rice as a model for comparative genomics of plants. Annu Rev Plant Biol 53:399–419. doi:10.1146/annurev.arplant.53.092401.134447
Song R, Llaca V, Messing J (2002) Mosaic organization of orthologous sequences in grass genomes. Genome Res 12:1549–1555. doi:10.1101/gr.268302
Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin MA, Ma X, Gustafson PJ et al (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Spielmeyer W, Singh RP, McFadden H, Wellings CR, Huerta-Espino J, Kong X, Appels R, Lagudah ES (2008) Fine scale genetic and physical mapping using interstitial deletion mutants of Lr34/Yr18: a disease resistance locus effective against multiple pathogens in wheat. Theor Appl Genet 116:481–490. doi:10.1007/s00122-007-0684-0
Vicient CM, Suoniemi A, Anamthawat-Jonsson K, Tanskanen J, Beharav A, Nevo E, Schulman AH (1999) Retrotransposon BARE-1 and its role in genome evolution in the genus Hordeum. Plant Cell 11:1769–1784
Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D (2006) EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor Appl Genet 113:186–195. doi:10.1007/s00122-006-0285-3
Wei F, Coe E, Nelson W, Bharti AK, Engler F, Butler E, Kim H, Goicoechea JL, Chen M, Lee S, Fuks G et al (2007) Physical and genetic structure of the maize genome reflects its complex evolutionary history. PLoS Genet 3:e123. doi:10.1371/journal.pgen.0030123
Wu J, Maehara T, Shimokawa T, Yamamoto S, Harada C, Takazaki Y, Ono N, Mukai Y, Koike K, Yazaki J et al (2002) A comprehensive rice transcript map containing 6591 expressed sequence tag sites. Plant Cell 14:525–535. doi:10.1105/tpc.010274
Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303:1640–1644. doi:10.1126/science.1094305
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586. doi:10.1073/pnas.0607142103
Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38. doi:10.1371/journal.pbio.0030038
Zhao S, Shatsman S, Ayodeji B, Geer K, Tsegaye G, Krol M, Gebregeorgis E, Shvartsbeyn A, Russell D, Overton L et al (2001) Mouse BAC ends quality assessment and sequence analyses. Genome Res 11:1736–1745. doi:10.1101/gr.179201
Zhao S, Shetty J, Hou L, Delcher A, Zhu B, Osoegawa K, de Jong P, Nierman WC, Strausberg RL, Fraser CM (2004) Human, mouse, and rat genome large-scale rearrangements: stability versus speciation. Genome Res 14:1851–1860. doi:10.1101/gr.2663304
Acknowledgements
The authors thank Roger Thilmony and William Belknap for the critical reading of this manuscript. This work was supported in part by the United State Department of Agriculture, Agriculture Research Service CRIS 532502100-000 532502100-011, and 532521000-13.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Huo, N., Vogel, J.P., Lazo, G.R. et al. Structural characterization of Brachypodium genome and its syntenic relationship with rice and wheat. Plant Mol Biol 70, 47–61 (2009). https://doi.org/10.1007/s11103-009-9456-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-009-9456-3