Abstract
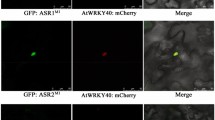
Anthocyanins and tannins are two of the most abundant flavonoids found in grapevine, and their synthesis is derived from the phenylpropanoid pathway. As described for model species such as Arabidopsis thaliana, maize and petunia, the end-point branches of this pathway are tightly regulated by the combinatorial interaction of three families of regulatory factors; MYB, bHLH (also known as MYC) and WDR proteins. Among these, only MYB genes have been previously identified in grapes. Here, we report the isolation of the first members from the WDR and bHLH families found in Vitis vinifera, named WDR1, WDR2 and MYCA1. WDR1 contributed positively to the accumulation of anthocyanins when it was overexpressed in A. thaliana, although it was not possible to determine the function of WDR2 by ectopic expression. The sub-cellular localizations of WDR1 and MYCA1 were observed by means of GFP-fusion proteins, indicating both cytoplasm and nuclear localization, in contrast to the localization of a MYB factor exclusively in the nucleus. The expression patterns of these genes were quantified in coloured reproductive organs throughout development, and correlated with anthocyanin accumulation and the expression profiles of the flavonoid-related MYBA1-2, UFGT, and ANR genes. In vitro grapevine plantlets grown under high salt concentrations showed a cultivar-dependent response for anthocyanin accumulation, which correlated with the expression of MYBA1-2, MYCA1 and WDR1 genes. These results suggest that MYCA1 may regulate ANR and UFGT and that this last control is easier to distinguish whenever MYBA genes are absent or in low abundance. Future studies should address the specific interactions of these proteins and their quantitative contribution to flavonoid synthesis in grape berries.






Similar content being viewed by others
Abbreviations
- bHLH:
-
Basic helix-loop-helix protein
- WDR:
-
Tryptophan-Aspartic acid repeat protein
- DFR:
-
Dihydroflavonol reductase
- UFGT:
-
UDP-glucose:flavonoid 3-O-glucosyltransferase
- PA:
-
Proanthocyanidin
- ANR:
-
Anthocyanidin reductase
- LAR:
-
Leucoanthocyanidin reductase
- GFP:
-
Green fluorescent protein
- WAA:
-
Weeks from/after anthesis
- WAV:
-
Weeks after veraison
References
Allan AC, Hellens RP, Laing WA (2008) MYB transcription factors that colour our fruit. Trends Plant Sci 13:99–102
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T (2007) Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol 48:958–970
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant Journal 39:366–380
Baudry A, Caboche M, Lepiniec L (2006) TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant Journal 46:768–779
Baxter N, Lilley TH, Haslam E, Williamson M (1997) Multiple interactions between polyphenols and a salivary proline-rich protein repeat result in complexation and precipitation. Biochemistry 36:5566–5577
Bogs J, Downey M, Harvey J, Ashton A, Tanner G, Robinson S (2005) Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol 139:652–663
Bogs J, Jaffe FW, Takos AM, Walker AR, Robinson SP (2007) The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol 143:1347–1361
Boss PK, Davies C, Robinson SP (1996) Analysis of the expression of anthocyanin pathway genes in developing Vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol 111:1059–1066
Carey CC, Strahle JT, Selinger DA, Chandler VL (2004) Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16:450–464
Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C (2008) Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 165:886–894
Cramer GR, Ergül A, Grimplet J, Tillett RL, Tattersall EA, Bohlman MC, Vincent D, Sonderegger J, Evans J, Osborne C, Quilici D, Schlauch KA, Schooley DA, Cushman JC (2007) Water and salinity stress in grapevines: early and late changes in transcript and metabolite profiles. Funct Integr Genomics 7:111–134
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
Czemmel S, Stracke R, Weisshaar B, Cordon N, Harris NN, Walker AR, Robinson SP, Bogs J (2009) The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. doi: 10.1104/pp.109.142059
de Vetten N, Quattrocchio F, Mol J, Koes R (1997) The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev 1:1422–1434
Deluc L, Barrieu F, Marchive C, Lauvergeat V, Decendit A, Richard T, Carde JP, Mérillon JM, Hamdi S (2006) Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol 140:499–511
Deluc L, Bogs J, Walker A, Ferrier T, Decendit A, Merillon JM, Robinson S, Barrieu F (2008) The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. doi:10.1104/pp.108.118919
Downey M, Harvey J, Robinson S (2003) Synthesis of flavonols and expression of flavonol synthase genes in the developing grape berries of Shiraz and Chardonnay (Vitis vinifera L.). Aust J Grape Wine Res 9:110–121
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC (2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49:414–427
Fujita A, Soma N, Goto-Yamamoto N, Shindo H, Kakuta T, Koizumi T, Hashizume K (2005) Anthocyanidin reductase gene expression and accumulation of Flavan-3-ols in Grape Berry. Am J Enol Vitic 56:336–342
Gollop R, Even S, Colova-Tsolova V, Perl A (2002) Expression of the grape dihydroflavonol reductase gene and analysis of its promoter region. J Exp Bot 53:1397–1409
Hartmann U, Sagasser M, Mehrtens F, Stracke R, Weisshaar B (2005) Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol Biol 57:155–171
Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20:735–747
Humphries JA, Walker AR, Timmis JN, Orford SJ (2005) Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol Biol 57:67–81
Jaillon O et al (2007) The French Italian public consortium for grapevine genome characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449:463–467
Jeong ST, Goto-Yamamoto N, Kobayashi S, Esaka M (2004) Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci 167:247–252
Kennedy JA, Matthews MA, Waterhouse AL (2000) Changes in grape seed polyphenols during fruit ripening. Phytochemistry 55:77–85
Kobayashi S, Ishimaru M, Hiraoka K, Honda C (2002) Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215:924–933
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Lin J-K, Weng M-S (2006) Flavonoids as Nutraceuticals. In: Grotewold E (ed) The Science of flavonoids, vol 8. Springer Science + Business Media, New York, pp 97–122
Liu Y-G, Huang N (1998) Efficient amplification of insert end sequences from bacterial artificial chromosome clones by thermal asymmetric interlaced PCR. Plant Mol Biol Rep 16:175–181
Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E (1991) Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J 1:37–49
Matus JT, Aquea F, Arce-Johnson P (2008) Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol 8:83
Matus JT, Loyola R, Vega A, Peña-Neira A, Bordeu E, Arce-Johnson P, Alcalde JA (2009) Post-veraison sunlight exposure induces MYB-mediated transcriptional regulation of anthocyanin and flavonol synthesis in berry skins of Vitis vinifera. J Exp Bot 60:853–867
Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S (2006) Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol 47:457–470
Nakatsuka T, Haruta KS, Pitaksutheepong C, Abe Y, Kakizaki Y, Yamamoto K, Shimada N, Yamamura S, Nishihara M (2008) Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol 49:1818–1829
Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L (2000) The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12:1863–1878
Park KI, Choi JD, Hoshino A, Morita Y, Iida S (2004) An intragenic tandem duplication in a transcriptional regulatory gene for anthocyanin biosynthesis confers pale-colored flowers and seeds with fine spots in Ipomoea tricolor. Plant J 38:840–849
Park KI, Ishikawa N, Morita Y, Choi JD, Hoshino A, Iida S (2007) A bHLH regulatory gene in the common morning glory, Ipomoea purpurea, controls anthocyanin biosynthesis in flowers, proanthocyanidin and phytomelanin pigmentation in seeds, and seed trichome formation. Plant J 49:641–654
Pattanaik S, Xie CH, Yuan L (2008) The interaction domains of the plant Myc-like bHLH transcription factors can regulate the transactivation strength. Planta 227:707–715
Payne CT, Zhang F, Lloyd AM (2000) GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156:1349–1362
Poupin MJ, Federici F, Medina C, Matus JT, Timmermann T, Arce-Johnson P (2007) Isolation of the three grape sub-lineages of B-class MADS-box TM6, PISTILLATA and APETALA3 genes which are differentially expressed during flower and fruit development. Gene 404:10–24
Quattrocchio F, Wing JF, van der Woude K, Mol JN, Koes R (1998) Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J 13:475–488
Ramsay N, Glover B (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Ramsay N, Walker A, Mooney M, Gray JC (2003) Two basic-helix-loop-helix genes (MYC-146 and GL3) from Arabidopsis can activate anthocyanin biosynthesis in a white-flowered Matthiola incana mutant. Plant Mol Biol 52:679–688
Reid K, Olsson N, Schlosser J, Peng F, Lund S (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT–PCR during berry development. BMC Plant Biol 6:27
Rowan DD, Cao M, Lin-Wang K, Cooney JM, Jensen DJ, Austin PT, Hunt MB, Norling C, Hellens RP, Schaffer RJ, Allan AC (2009) Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol 182:102–115
Smith TF, Gaitatzes C, Saxena K, Neer EJ (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem Sci 24:181–185
Somers T, Evans M (1974) Wine quality: correlations with colour density and anthocyanin equilibria in a group of young red wines. J Sci Food Agric 25:1369–1379
Sompornpailin K, Makita Y, Yamazaki M, Saito K (2002) A WD-repeat-containing putative regulatory protein in anthocyanin biosynthesis in Perilla frutescens. Plant Mol Biol 50:485–495
Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50:660–677
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. doi:10.1093/molbev/msm092
Terrier N, Torregrosa L, Ageorges A, Vialet S, Verriès C, Cheynier V, Romieu C (2009) Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol 149:1028–1041
van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 12:50
Vidal S, Francis L, Guyot S, Marnet N, Kwiatkowski M, Gawel R, Cheynier V, Waters EJ (2003) The mouth-feel properties of grape and apple proanthocyanidins in a wine-like medium. J Sci Food Agric 83:564–573
Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC (1999) The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11:1337–1350
Walker AR, Lee E, Bogs J, McDavid DA, Thomas MR, Robinson SP (2007) White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant Journal 49:772–785
Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Acknowledgments
We wish to thank Uely Grossniklaus (University of Zurich, SZ) for the p207-DONOR and pMDC84 GFP-fusion vectors, and Paula Salinas and Dave Jackson (Cold Spring Harbour Laboratory, NY) for their guidance on GFP visualization in tobacco and onion cells, respectively. Special thanks to Agnès Ageorges and Nancy Terrier (INRA Montpellier) for sharing bHLH and WDR QPCR primers. This work was supported by the Chilean Wine Consortium 05CTE01-03, the Fruit Consortium, 07Genoma01, Millennium Nucleus for Plant Functional Genomics (P06-009-F), FONDECYT 1100709, and by fellowships awarded to JTM (MECESUP and CONICYT AT24060171) and MJP (CONICYT). Finally, we thank Nancy Terrier and Michael Handford for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
A. Alignment of deduced amino acid sequences of WDR1 and WDR2 and theirhomologues in plant species. The four repeats are boxed. B. Comparison between WDrepeats identified in WDR1 and WDR2 and the WD-repeat consensus sequencedescribed by Smith et al. (1999), which includes the a, b and c beta strands that form thebeta-propeller structure. Identical amino acid residues are shaded in yellow whilesimilar residues appear shaded in green or cyan. Dots represent gaps introduced toimprove the alignment (JPG 928 kb).
Figure S2
Best-fit alignment of the deduced amino acid sequences homologous to VvMYCA1.The MYC DNA-binding basic region is followed by two alpha-helices separated by avariable loop region. The positions of the interacting and bHLH domains are shown.Identical amino acid residues are shaded in yellow while similar residues appear shadedin green or cyan. Dots represent gaps introduced to improve the alignment (JPG 1027 kb).
Figure S3
Changes in berry weight (•), total soluble solids (º Brix) (□), and anthocyanins per gramfresh weight of berry skin (▲), measured at a two week interval during grapedevelopment (JPG 58 kb).
Figure S4
Affymetrix-based expression analyses of MYCA1,WDR1 and anthocyanin-relatedUFGT and MYBA genes in grapevine shoot tips under abiotic stress conditions. Days 8,12 and 16 after control, water stress or salt (NaCl) treatments are shown for each gene.Data taken from the grape Plant Expression (PLEX) database for microarray data(http://www.plexdb.org/plex.php?database=Grape) (JPG 140 kb).
Rights and permissions
About this article
Cite this article
Matus, J.T., Poupin, M.J., Cañón, P. et al. Isolation of WDR and bHLH genes related to flavonoid synthesis in grapevine (Vitis vinifera L.). Plant Mol Biol 72, 607–620 (2010). https://doi.org/10.1007/s11103-010-9597-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-010-9597-4